Genetics 2 - Chromosomes: Structure and Abnormalities Flashcards
(78 cards)
learning outcomes

chromosome structure
Chromatin = DNA and protein
Protein - histones - tightly packing
1m of DNA into each of cells
Important for structure and control of transcription of DNA

chromatin structure
Bead - nucleosome - 150bp
DNA wrapped around
Modification of histones influences transcription
- Phosphorylation
- Methylation
- acetylation

euchromatin
gene switched on
active (open) chromatin
unmethylated cytosines (white circles)
acetylated histones

heterochromatin
gene switched off
silent (condensed) chromatin
methylated cytosines (red circles)
deacetylated histones

cytogenetics
- sample nucleated cells you can grow easily - T cells/skin fibroblasts/bone marrow/chorionic villi or amniotic fluid (antenatal)
- get chromosomes into a form in which they can be observed and differentiated from each other (metaphase chromosomes - grow the cells to dividing stage)
- chemical preparation to arrest cells and lyse nuclei
- stain chromosomes (giemsa)

conventional cytogenetics
limitation
identify chromosomes by banding patterns
arrange homologs side by side
⇒ karyogram
described as a karyotype
46, XX female and 46, XY male
limitation = resolution
detects GROSS chr changes (> 5Mb)

international system for human cytogenetic nomenclature (ISCN)

molecular cytogenetics
MOA
what is it used for
probe hybridisation - visualisation of location of specific nucleotide sequence (< 5Mb)
USES
is sequence present and if so on which Chr
are 2 sequences close to each other
preimplantation genetic screening

Fluorescent In Situ Hybridisation

multiplex FISH (M-FISH)
metaphase chr
detects small Chr rearrangements
uses painting probes
paint whole Chr

structural chr abnormalities
categories
heritability
change in chr structure
caused by chr breakage with subsequent abnormal realignment
BALANCED: no gain/loss of genetic content ⇒ often not clinically relevant but relevant if they interrupt a gene with dominant loss of function
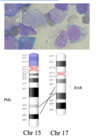
Or GOF via fusion of 2 unrelated segments - CML
UNBALANCED: gain or loss of genetic content ⇒ clinically relevant
structural Chr abnormalities are heritable only if in germ line

deletion
part of chr deleted
sever effects
large deletions are lethal
occurence of large deletions
example
> 5 Mb
visible under microscope (conventional cytogenetics)
e.g. crie-du-chat syndrome (5p)
smaller deletions occurence
how are they visualised
e.g.
microdeletions are not visible under light microscope
some visible with FISH
smaller ones using chr oligonucleotide arrays - microarrays
e.g. Smith-Magennis Syndrome del (17p11.2)
duplication
e.g.
chr section repeated (often sequentially)
less sever than deletion of Chr section
e.g. Potocki-Lupski syndrome dup(17p11.2)

copy number variants
Mbs
role in….
deletions/duplications of genomic sequence
1 Kb → Mbs (variable no of repeats of a particular unit of sequence)
major determinants of human genetic variation - 5-10% of genome is CNVs
can represent functional (disease causing) mutations as well as genomic polymorphisms of uncertain relevance
inversions
e.g.
2 breaks occur in chr
part of chr in between is rotated 180 degrees before re-joining
results in looped chr pairs in meiosis I
can cause recombinant chrs if cross over happens within loop
e.g. F8 inversions (intron 22) in haemophilia A

translocation
non-homologous chromosomes exchange segments
reciprocal translocation
2 chromosomes swap non-homologus sections
chr number and genetic content are unchanged
common

robertsonian translocation
break occurs at the short arm of acrocentric chromosomes (13, 14, 15, 21, 22, Y) and results in fusion chr
change in chr number (45) - short p arms lost

Down Syndrome - chr change
4% is translocation (familial) DS
rob(14;21) or rob(21;21) + 21
balanced translocations
no extra or missing material (reciprocal)
DNA just packaged incorrectly
phenotype usually normal
robertsonian translocations = balanced
robertsonian
balanced translocation