DNA to RNA to Protein COPY Flashcards
DNA replication enzymes
DNA replication enzymes form a large machine
Key proteins: helicase, SSB, primase, polymerase, ligase, and topoisomerase
Helicase unwinds DNA helix (needs ATP)
Single stranded binding protein (SSB) stabilizes single-stranded DNA
Primase synthesizes short complementary RNA primer (provides 3’ OH)
DNA polymerase synthesizes new DNA in a 5’ to 3’ direction ONLY
Ligase seals the break in DNA
Topoisomerase relieves twisting, turning, and knots in DNA (supercoiling)
single stranded binding protein
Single stranded binding protein (SSB) stabilizes single-stranded DNA
basically stabilizes single strands so do not tangle up, lots of thread all over the place need protein to prevent tying itself in knots**
Primer for DNA replication
Primase synthesizes short complementary RNA primer (provides 3’ OH)
RNA PRIMER than DNA polymerase adds onto primer
RNA primer means that stretch of RNA has to later be replaced by dna
for dna replication….
what kind of bonds are we breaking?
when breaking strands, breaking hydrogen bonds btwn two strands of dna helix
when use ligase to join together okazaki fragments, connecting parts of sugar phosphate backbone those are covalent bonds called phosphodiesterase bonds*; covalent bonds of sugar phosphate backbone
sugar phosphate sugar phosphate and bases connect to eachother in the middle*
middle image, pannel b if on edges 5’ end and blue sugar, when reffering to phosphate backbone is the blue, those bonds from one nucletide to the next are covalent bonds called phosphodiesterase bonds, what ligase catalyzes, puts together okazaki fragments through new phosphodiester bond* middle of DNA helix where bases connect to eachother and conenct through H bonds, A and T and G and C two strands connected down the middle with hydrogen bonds
Ligase
connects Okazaki fragments on lagging strand, makes covalent bonds
each strand is covalently bonded= phosphodiesterase bonds*
covalent bonds of sugar phosphate backbone
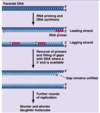
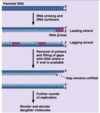
DNA replication mechanism
Enzymes first assemble at replication origin
Helicase → SSB → primase → DNA pol
Replication fork (Y-shaped junction) moves as DNA is replicated
Both new daughter strands are synthesized at the fork
Leading and Lagging strands
Leading strand synthesis is continuous (direction of replication fork movement)
Lagging strand synthesis is discontinuous (away from replication fork)
On lagging strand, a new RNA primer each ~200 bases in eukaryotes
Lagging strand DNA fragments = Okazaki fragments
RNA primer replaced with DNA by polymerase
Ligase seals the break in DNA between Okazaki fragments
Topoisomerase relieves twisting, turning, and knots in helix
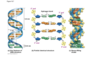
DNA structure 1
DNA is a nucleotide polymer that underlies heredity
Usually double stranded
Each nucleotide has a pentose sugar, phosphate, and nitrogenous base
Alternating sugar and phosphate form helix backbone
Phosphates give DNA a negative charge
Bases are on the interior of the helix
DNA structure 2
Four nucleotides: adenine, guanine, thymine, cytosine (A, G, T, C)
Purines: A and G (two rings)
Pyrimidines: C and T (single ring)
Mnemonic: on a $1 bill, the pyramid is CUT
A pairs with T, two hydrogen bonds
G pairs with C, three hydrogen bonds
Amount of A = T, G = C in double stranded DNA (Chargaff ’s rule)
The two strands of the double helix are antiparallel (one 5’ to 3’, one 3’ to 5’)
Each turn (360°) = ten base pairs
Nucleoside = pentose sugar and nitrogenous base but no phosphate
Chargaff’s rule
Amount of A = T, G = C in double stranded DNA (Chargaff ’s rule)
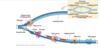
image description of DNA replication
ALWAYS added onto 3’ side, so on leading strand going into replication fork continuously, as zipper unzips that leading strand can just go go go
whereas on the lagging strand, the synthesis of DNA is discontinuous
so first of okazki fragments, so earlier stages red represents primer, dna polymerase making laggings trand and then DNA polymerase I replaces rna primer I with DNA, then DNA ligase makes phosphdiester bond that puts together fragmetns
DNA polymerase III is the work horse, used for most of the synthesis of DNA layign down of new DNA is done by DNA poly. III
DNA poly. I is specially equipped to replace the RNA primer with the equivalent DNA letters
Fidelity of DNA replication and proofreading
Errors are made spontaneously during DNA replication
Error rate is remarkably low (1 in 109)
“Proofreading” reduces errors and increases replication fidelity
DNA polymerase examines what it just synthesized
If pairing is wrong, polymerase pauses, removes mismatched base, resumes synthesis
Proofreading reflects exonuclease activity of polymerase (moves backwards, 3’ to 5’)
DNA polymerase can go backand cut out the wrong letter and replcae it with the correct letter, called an exonuclease function cutting out last letter put in
mutations and dna repair
Mutations occurs spontaneously, from replication errors, UV, X-rays, chemical mutagens
Spontaneous deamination (C → U), (methyl-C → T)
Spontaneous depurination (A and G base can be lost)
UV can crosslink T dimers
X-rays can cause double-stranded breaks
Some chemical mutagens increase rate of replication errors
Repair of DNA is facilitated by having two strands in helix
mutations and dna repair 2
- UV light can cross link thymine dimers, causing two thymies next to eachother on same strand to form extra bonds with eacother form a dimer and create a bulge in dna strand*** the way uv light is the road ot skin cancer** how UV light damages dna
Xrays are really bad news cause double stranded breaks, harder to repair
environmental chemicals, stuff we are exposed to can cause whole range of errors
Spontaneous deamination (C → U), (methyl-C → T), if look at chemical structures of letters sometimes turning one into another, if mess with one substituent on ring have different nitrogenous base, if alter the chmeistry of one base can turn it into another base
- Deamination literally means taking off the NH2 group and replacing it with a o double bonded

DNA repair mechanisms
Cells monitor DNA damage constantly and repair DNA
Cells have checkpoints to delay replication until damage is fixed
- Mismatch repair fixes mismatched base pairs after replication
- Base excision repair repairs one nucleotide (e.g., from deamination)
- Nucleotide excision repair fixes bulkier damage (e.g., crosslinked T dimers from UV)
- Double-strand break repair fixes a complete break in the double helix
general idea is so many enzymes devoted to repairing errors in DNA mutations or damage, and you can sort of think about them in terms of how big of a problem they address
ex. base excision reapir cuts out one and put right one in
nucletoide excision repair- bulkier damage, cuts out 10-20 so its like hwo big is the job have differnet enzymes etc.
if damage on one strand and enzyme come sina nd takes out huge chunk can often have complete repair, if just one strand has to be cut out then still have other strand which is complementary/record of what the right letters to go in should be
vs. Double strand break repair is what happens when have a complete break through both strands of DNA, what kind of damage get from xrays and that is somewhat salvagable but not completely issue is you are trying to glue together broken ends but usually lose some dna letters around the ends when you do that, there is a fix but it is not a perfect fix**
If you have mutations with DNA repair systems, you can really really have problems*
ribozymes
Can catalyze reactions, so supports can have reproducing primitive bits of RNA considered earliest lief forms- rna world
A few RNAs can acts as enzymes
RNA enzyme = ribozyme
Example: self-splicing intron
Catalytic ability of RNA supports RNA world hypothesis (RNA came first)
snRNPs
snRNPs = small nuclear RiboNucleoProtein complexes
snRNPs contain snRNAs (small nuclear RNAs)
mash up of rna and protein that do splicing* sp evidence of rna serving a catalytic role* rna world, point is its not just protein that catalyzes spelicing there is some rna in there also
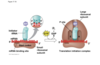
initation of translation
mRNA has on it start codon* AUG and hte first trna carrying the met attaches htere, 3’ UAC 5’
then large ribosomal subunit comes on top, everythign comes together around tnra and mrna
mrna what we just made through transcription, if go along mrna 5’ to 3’ three letters at a time, starting with AUG those are the codons
so codons are made of mrna, and the codons code for amino acids
trna= actually bring over hte amino acids needed according to code, tRNA has the anticodon of hte codon on mrna, so the compliment!
anticodon is complimentary to codon, but matching up of letters is also antiparalllel** codon 5’ to 3’ AUG, but anticodon is 3’ to 5’, 5; end of anticodon above 3’ end of codon, so sometimes they will say which anticodon goes with aug* answer choices will all be written 5’ to 3’, they would label it but correct answer to what anticodon is aug is CAU**** can seem wrong because its backwards** be very careful which is 5’ ends and 3’ end
first trna is in P site
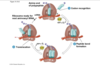
translation elognation
- new trna coming into A site, A for amino acid, next for amino acid comes into that site
- peptide bond formation, whole polypeptide chain in p site jumps over on top of new guy
- 6 oclock, whole chain now held on top of trna at A site temporarily, but then you need step three translocation where everything slides over
- 9 oclock, empty trna gets kicked out of exit site, to go pick up amino acid again for future; meanwhil in P site trna holding growing polypeptide chain, A site now empty so can now go back aroudn to 12 oclock and another trna with another amino acid can come in and keep elongation process going
translation: termination
UGA, UAA, UAG
You go away, You are away, You are gone
**stop codons can be called amber* an amber codon is the same thing as a stop codon, weird terminology they love***
so when get to a stop codon, no trna but protein called release factor that binds to stop codon and causes the release of the polypeptide, trna and mrna, everythign is released and ribosome comes apart

energy for translation…..
KNOW FOR TRANSLATION GTP, translocation involves GTP, final step of termination invovles GTP*** remember which steps require energy

proteins are synthesized ….
N to C****
first amino acids to come off hte ribosome or the end that is made first is the amino end***
as each additional amino acid is beign added hte synthesis is proceeding toward carboxy end of peptide**
so if you have a problem, some mutaiton related problem in middle, amino end may be fine, like if shift in reading frame screw with entire amino acid, more liekly to have problem going toward carboxy, carboxy side is more screwed up because made later*
Cytoplasmic vs secreted and transmembrane protiens
- Cytoplasmic proteins are translated on free ribosomes
- Secreted and transmembrane proteins are translated on rough ER
- Secreted proteins and transmembrane proteins have signal peptide
- Signal peptide causes ribosome to pause translation, dock on rough ER
- Translation resumes, protein inserted into rough ER during translation
- Signal peptide is subsequently cleaved off protein
signal peptide= mailing label, part of amino acid sequence, series of amino acid at the terminus, but cell responds to it as a diretive to send the growing peptide to the membrane of the ER so peptide can go into ER and then proceed through that endo membrane system and then end up three fates: exocytosis from cell, inserted into plasma membrane or sent to a lysozome*
Post-translational modification
Once proteins leave the ribosome they can be modified further
Modifications can affect protein activity and function
Disulfide bonds for extracellular proteins (S-S bonds formed by cysteines)
Phosphorylation can regulate activity
Kinases add P
Phosphatases remove P
Glycosylation (add sugars in ER and Golgi)= after protein itself is made all sorts of ways like adding sugar groups, proteins can be modified
Proteolytic processing (e.g., zymogens)

Amino acid configuration
Have absolute configuration of D or L at the alpha carbon
D = amino group is on right when COOH is up and R is down in Fischer projection
L = amino group on left when COOH is up and R is down in Fischer projection
Amino acids in body are L
Enzymes in the body can be inhibited by D amino acids, which can bind to active site but not react
Protein structure
Four levels of protein structure
- Primary (1°) structure = amino acid sequence of a polypeptide chain
- Secondary (2°) structure = alpha helices and beta strands/sheets
- Involves hydrogen bonding between backbone atoms
- Tertiary (3°) structure = globular 3-dimensional fold of a single polypeptide
- Involves interaction between amino acid side chains
- Can include hydrophobic-hydrophobic and hydrophilic-hydrophilic interactions
- Can include hydrogen bonds and ionic bonds
- Can include disulfide bridges = covalent bonds between cysteine residues (S—S)
- It is favorable for hydrophobic side chains to be buried in protein, away from aqueous environment = lower “entropic penalty”
- Quaternary (4°) structure = assembly of several polypeptide chains
in the body, amino acids are….
L
if active site wants to work with L, most of what is in the body is L; intervention to inhibit that enzyme one thing can do is throw in D substrate to get into active site, be a compeitive inhibitor since stereospecific for amino acids, if threw in D amino acids can act as inhibitor of enzyme, so remember enzymes fuzzy when comes to working with L amino acids
wrong enamtioner- D will bidn but reaction will nto happen, so great competitive hinhbitior for nezymes that build things up to bigger things or use for various purposes in the cell
L= natural substrates
D= inhibitors

three amino acids show up in UV
In UV spectroscopy, tryptophan, phenylalanine, and tyrosine have distinctive absorptions (around 270nm) because of their aromatic rings
can use to see presence of peptide, some big protein will have some of these aromatic amino acids
KNOW 270 NM*
coding sequence mutations 1
mutaiton in exon gives you a nonfunctonal protein but other mutations that can effect other aspects of the process, love to give you a scenario and ask where do you think mutaiton was
ex. too much of a protein, but activity is normal still doing what it is suppsoed to do but jsut way too much of it, mutaiton in promoter or enhancer some issue with regulation
ex. introns or exons, if something happens introns stay in when it is supposed to get cut out with splicing, mutaitions in splice sites meaning abnormal splicing*** if mutation in splice site donor site or acceptor site, all this specificty you do not need to know, speculating about what a possible mutaton may be- go for any answer choice seems plausible if cant arrive there on your own, rule out ones that are wrong ok to chose something more specific then necessarily known on my own but nothing wrong with it
coding sequence mutaitons 2
Point mutation = a single base pair mutation
Four classes of coding sequence mutations: silent, missense, nonsense, and frameshift
Silent mutation causes no change in amino acid sequence (e.g., third codon position)
Missense mutation changes one amino acid in the protein
Nonsense mutation introduces a premature STOP codon
Frameshift mutation results from the insertion/deletion of bases in non-multiples of three
Frameshift alters the reading frame and protein sequence downstream of the mutation
Frameshift usually brings a new STOP codon into frame as well
point base mutation
Point mutation = a single base pair mutation
one letter, one little tiny point in dna is messed with
nonsense mutation
Nonsense mutation introduces a premature STOP codon
just has one of three stop codons there, it turns something that was supposed to code for an amino acid to UAG, UAA, UGA
silent mutations
No change in amino acid becuase of redundancy of code
Silent mutation causes no change in amino acid sequence (e.g., third codon position)
missense
missense- one amino acid different, one letter can be different if change one letter of codon can be coding for a diff amino acid
Missense mutation changes one amino acid in the protein
Frameshift
Can be one or more just not multiples of 3, all very small scale
Frameshift mutation results from the insertion/deletion of bases in non-multiples of three
Frameshift alters the reading frame and protein sequence downstream of the mutation
Frameshift usually brings a new STOP codon into frame as well
other mutations that affect gene expression
Mutations can affect all levels of gene expression:
Can affect gene transcription (e.g., mutation in promoter or enhancer region)
Can affect splicing, mRNA stability, or translation
Can affect protein level, stability, or activity
Chromosomal mutaitons 1
Can insert a huge chunk/stretch of DNA being inserted like from a virus for instance
duplication- two copies of the same gene, retain both, can be a big deal in terms of evolution of proteins how can get family of proteins, imagine you have a gene for a protien like transporter then duplicate gene, first copy can still pump put original protein serve needs of cell, second copy could also do that but also immitate and have some mutations to be a different version of the proteins; allows hte cell to innovate and have similar proteins evolving, how you get a whole family of proteins or calcium transproters, whole family have related functions but do it in slightly different ways
insertions
a chromosomal mutation
Insertions: Can involve addition of larger segments of DNA into chromosome
DNA can come from another chromosome (e.g., from errors in crossing over)
Can come from another source, like virus
Can disrupt enhancers, promoters or genes themselves
inversion
(a chromosomal mutation)
Inversions: involve “flipping” a DNA sequence within its chromosomal location
• Can cause loss of function or potentially gain of new function
a chromosomal mutation
Deletions
chromosomal mutation
Deletions: can involve removal of larger DNA sequence
translocations
chromosomal mutaitons
Translocations: involve moving a sequence from one chromosome to another
Translocations chunk of one chromosome moves over and attaches to a different chromosome, lots have to do with cancer, disrupt gene has to do with cell growth get cell growth happening in an out of cotnrol way
Transposons jumping genes, similarly piece of dna jumping to a new location and new piece of dna inserting itself, jumping over that can jump into middle of another gene and mess with that gene, jump into middle of promoter or enhancer can mess with regulation fo another gene; very disruptive
methylation vs acetlation
methylation suppresses gene expression
acetylation inhances gene expression
heterochromatin
very very tighlty packed dna that will not be transcribed
barr bodies, one X randomly condensed into a barr body true in each skin cell everything except from germ cells, examples of whoel X chromsome in females in each cell being condensed, centromeres and telomeres are also very very tightly packed so made of heterochromatin
goes alogn with fact no genes being transcribed so you cannot transcribe heterochromatin becuase it is too tightly packed for rna polymerase to access, but there is nothing on centromere worth transcribing, centromeres are there to help with mitosis* so similarity with telomeres there are no genes being transcribed from telomeres
telomeres
- thing with linear chromosome, ends of hte chromosome
- everytime cell replicates or divide telomeres get shorter
- point of image is to show why that is= can see how RNA primer what this image is tryingn to show is that there is no way to replace that RNA primer, it gets removed but dna polymerase cannot parachute in from right there and fill in gap, DNA polymerase has to always work off of a preexisting nucleotide with 3’ OH so gap that remains unfilled, as replication occurs daughter molecules of DNA keep getting shorter becuase of that issue with not quite able to fill in where primer was on very end of the strand
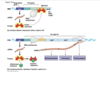
telomeres 2
Telomeres = heterochromatin
Telomere = stretch of repeated DNA + bound proteins
Telomere repeats are synthesized by telomerase enzyme
Telomeres 3
On linear chromosome, cannot replicate lagging strand all the way to the end
Lagging strand gets shorter with each replication
Telomere “caps” the end of the chromosome and counteracts shortening
Telomeres = heterochromatin
Telomere = stretch of repeated DNA + bound proteins
Telomere repeats are synthesized by telomerase enzyme
Telomerase contains an RNA molecule and is a reverse transcriptase (RT)
Telomerase copies RNA piece into chromosome → new DNA repeat
Loss of telomeres may cause senescence (no more cell division)
why gap, where RNA primer is at end
why telomeres shorten 2
- there is no way to replace that DNA polymerase cannot get around to the next side, the next cannot hook on there because 5’ end, dna polymerase can only extend 3’ end so if look at this gap but dna polymerase cannot do anything here becuase would have to be going the opposite way, but that is the wrong direction* so there is a gap
- with each generation of cells and replication of DNA you lose more of the telomeres, we have evolved these telomeres are random letters tehre to be degraded, letters are not important
- just put a limit in how many times cells can divide, not infinaity long, so why after 30 rounds of division most cells go into period of dormance beucase ran out of telomere to keep going to dont keep dividing
- need stem cells to keep dividing and diviiding so thye are able to turn on the gene for an enzyme called telomerase, which is able to bring a special process to extend telomeres; telomerse not activated in most cells, only stem cells and cancer cells, cancer cells immortal just divide adn divide and divide, natural stop to that telomeres would normally force a cell to stop dividing but cancer cells activate telomerase which makes telomeres grow back and then cancer cells can keep going and dividing more
- as age or go through periods of stress that shortens the telomerase, many ppl trying to market telomere tests to see internal biological age

operons
In prokaryotes, several genes typically transcribed together = operon
ONLY IN PROKARYOTES
operon 2
- promoter region, next to operator, then genes
- if repressor bound to operator, rna polymerase (white blob) cannot get through and reach genes
- when lactose is present, lactose binds to repressor, repressor comes off operator and transcription occurs make these three enzymes shown at the bottom
- also ex of prokaryotes being polysincronic, becuase from one stretch of mrna making three proteins
- inducible operon, default state off
- when certain moelcule, lactose present then transcribe the genes for enzymes that break down lactose
- tryp operon is repressible, opposite where default state of transcirption is on, when tryptophan is present the repressor binds and that stops production*
- PROKARYOTES
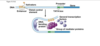
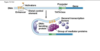
EU role of transcription factors…
binding to enhancers and promoters, no operon
ton of proteins getting in there binding to promoter and enhancers which can be upstream of a gene or downstream of a gene, can be far away
get invovled directly from DNA binding around on itself, loop around together to enable transcrtiption
we use transcription factors, all kinds of complex transcription factors binding! why for prokaryotes when think of operon genes grouped together in operon are genes want to express at the same time, under same promoter simple way of doign it; for us want genes expresed at same time but on totally different chromosomes, so need complex transcirption factors to regulate it and do it
enhancers made of DNA part of DNA< when certain transcription factors bind to enahncers that activates them and they have a say in gene expression; different transcription factors are present in differnet types of cells, important stretch of DNA in brain cells improtant certain genes activated in brain but nothing binds to it in stomach cell, so some just dormant in stomach cell, so different combinations of transcription factors binding to ehancers how get specific tissue expressions of gene expression, also how get specific gene expression in different stages of development, some transcription factors very important in early stages of embryo and then other transcription factors when have more differentiated cells, waves of transcription factors important to development
Transcription
- Transcription is the act of making a complementary RNA copy from DNA
- In eukaryotes, one mRNA encodes one protein (monocistronic)
- In prokaryotes, one mRNA can encode multiple proteins (polycistronic)
- RNA polymerase synthesizes RNA, in a 5’ to 3’ direction ONLY
- The antisense strand is transcribed = template strand
- Polymerase binds at the promoter of a gene
- Promoter tells RNA polymerase where to begin transcription
- A terminator tells RNA polymerase where to stop transcription
- Prokaryotes have one RNA polymerase
- Eukaryotes have three RNA polymerases
- RNA Pol I synthesizes most rRNA
- RNA Pol II synthesizes mRNA
- RNA Pol III synthesizes tRNA, some rRNA
DNA replication
Replication is semi-conservative: each parent strand is a template for a new strand
Each DNA helix has one old strand and one newly synthesized strand
DNA replication begins at a specific site called a replication origin
Prokaryotes have one replication origin
Eukaryotes have many replication origins
mature RNA
(mRNA)
In eukaryotes, pre-mRNA → mRNA: (1) capping, (2) splicing, (3) polyA tail
5’ end of message is capped with modified guanine nucleotide
Capping necessary for ribosomal binding and translation
3’ end of message is polyadenylated (polyA tail is 100–250 bases)
polyA increases mRNA stability
Eukaryotic genes have introns (intervening sequence)
Introns are spliced out of pre-mRNA
Mature mRNA contains only exons (expressed sequences)
Helicase
Helicase unwinds DNA helix (needs ATP)
Primase
Primase synthesizes short complementary RNA primer (provides 3’ OH)
DNA polymerase I and III
DNA polymerase synthesizes new DNA in a 5’ to 3’ direction ONLY
III is the main guy, workhorse
Ligase
Ligase seals the break in DNA
topoisomerase
Topoisomerase relieves twisting, turning, and knots in DNA (supercoiling)
mRNA maturation and splicing
Spliceosome = machinery of splicing
Spliceosome = snRNPs plus additional proteins
snRNPs = small nuclear RiboNucleoProtein complexes
snRNPs contain snRNAs (small nuclear RNAs)
Eukaryotic mRNAs can be alternatively spliced (e.g., exons 12345 or 1245 or 135)
Alternative splicing can produce different proteins from one gene
Final mRNA structure:
5’ untranslated leader sequence with ribosome binding site
Coding region is translated to protein, defined by START and STOP codons
3’ untranslated sequence follows coding sequence
DNA repair mechanisms 2
mismatch repair
- Mismatch repair fixes mismatched base pairs after replication
- Old strand is methylated, newly replicated strand is not
- Mismatch repair mechanism sees “bulges” from incorrect pairing
- Error is removed from new (unmethylated) strand, DNA polymerase & ligase fix
Base excision repair
Dna repair mechanisms
Base excision repair repairs one nucleotide (e.g., from deamination)
• Base is cut out, replaced by DNA polymerase, and ligase seals helix
Nucleotide excision
DNA repair mechanisms
Nucleotide excision repair fixes bulkier damage (e.g., crosslinked T dimers from UV)
- 10–20 DNA bases around damaged region are excised on one strand and replaced
Doube-strand break repair
Double-strand break repair fixes a complete break in the double helix
- Broken ends can be glued together, but some sequence is lost
- Sometimes, homologous DNA can serve as template to restore missing DNA
DNA repair and cancer
Defects in DNA repair lead to accumulation of mutations, increased cancer
Example: xeroderma pigmentosum (XP) results from defective excision repair
XP individuals are ultra-sensitive to UV, predisposed to skin cancer
RNA structure
RNA uses ribose sugar (has extra 2’ OH) instead of deoxyribose
Usually single stranded
Uracil (U) instead of (T); U pairs with A
messenger RNA (mRNA) encodes a protein
transfer RNA (tRNA) decodes mRNA during translation
tRNA can sometimes contain modified nucleotides (beyond just A, U, C and G)
ribosomal RNA (rRNA) is a key part of the ribosome
Final mRNA structure:
5’ untranslated leader sequence with ribosome binding site
Coding region is translated to protein, defined by START and STOP codons
3’ untranslated sequence follows coding sequence
the genetic code
The genetic code dictates how mRNA is translated to protein
Universal and shared by all organisms
Based on nucleotide triplet codons (3 nucleotides → one amino acid)
Non-overlapping: three bases, then the next three
Degenerate: 43 = 64 codons, but only 20 amino acids (>1 codon per amino acid)
For a single amino acid, codons generally vary at third position (wobble)
Cognate codons = code for the same amino acid
Codons in mRNA are recognized by complementary anticodons in tRNA
translation 1
Translation is the act of generating protein from an mRNA
Occurs in the ribosome
Ribosome composed of many proteins and rRNA
Translation requires tRNAs
tRNAs are “charged” at 3’ end with single amino acids (tRNA-AA)
Aminoacyl synthetase enzymes charge tRNAs
mechanism of translation
Ribosome has 2 sites for tRNA binding: P and A sites
P (peptidyl-tRNA site) holds growing protein
A (aminoacyl-tRNA site) holds tRNA-AA
Small ribosome subunit binds mRNA, slides to START codon, tRNA-MET binds in P
Start codon = AUG
Large subunit joins assembly
2nd tRNA-AA sits down in A and pairs with next codon
Peptide bond forms, releases 1st tRNA which leave ribosome
2nd tRNA-dipeptide is still bound in P
Ribosome moves 3’ to next codon, 2nd tRNA-dipeptide now in A
Cycle repeats until STOP codon (UGA, UAA, or UAG)
STOP codon releases protein and ribosome disassembles
5’ to 3’ translation makes protein in an amino to carboxy (N to C) direction
tertiary structure of proteins
Tertiary (3°) structure = globular 3-dimensional fold of a single polypeptide
Involves interaction between amino acid side chains
Can include hydrophobic-hydrophobic and hydrophilic-hydrophilic interactions
Can include hydrogen bonds and ionic bonds
Can include disulfide bridges = covalent bonds between cysteine residues (S—S)
It is favorable for hydrophobic side chains to be buried in protein, away from aqueous environment = lower “entropic penalty”
hydrophobic amino acids
Alanine
Valine
Leucine
Isoleucine
Proline
Phenylalanine
Methionine
Glycine
Cysteine
Tryptophan
polar uncharged
Asparagine
Glutamine
Tyrosine (-OH)*
Threonine (-OH)*
Serine (-OH)*
these three * can be phosphorylated
negative amino acids
Asparate
Glutamate
positively charged amino acids
Lysine
Arginine
Histidine
glycine
- The side chain is –H, which allows for greater flexibility in protein structure
- Glycines tend to appear as “hinges” in alpha helices
Proline
- Proline has a secondary amino group (amino N bonded to two carbons)
- Proline constrains protein conformation and often creates “kinks”
Cysteine
Cysteine contains sulfhydryl (-SH) groups in its side chain
Can be oxidized to form covalent S-S cross bridges (disulfide bonds) with other cysteines
Disulfide bonds tend to constrain protein structure and add stability
Disulfide bonds are more common in secreted proteins that need added strength
Disulfide bonds also tend to be unstable in the cytosol, which is a reducing environment
Tyrosine, Serine, and threonine
Tyrosine, threonine, and serine have –OH’s on their side chains
They can be phosphorylated by protein kinases (then dephosphorylated by phosphatases)
Tyrosine is the starting material for several neurotransmitters and hormones
Examples = dopamine, norepinephrine, epinephrine, and T3 and T4 (thyroid hormones)
Tryptophan
Tryptophan is the starting material for the neurotransmitter serotonin
REMEMBER NOT PHOSPHORLATED, not tyrosine or threonine*
glutamate
Glutamate serves as the primary excitatory neurotransmitter in the central nervous system
amino acids, ph, pka
pKa = pH at which group is half protonated and half deprotonated
Example: the side chain of lysine has a pKa of 10.5. For 2mM of lysine at pH =10.5, 1mM has side chain with –RNH3+ and 1mM has side chain with –RNH2
Cellular pH is roughly 7.4
Amino acids with basic side chains are predominantly protonated
Amino acids with acidic side chains are predominantly deprotonated
Examples: Glutamic acid and aspartic acid (pKa’s approx = 4), are mostly deprotonated
Called “glutamate” and “aspartate” when deprotonated
Amino acids can be separated in a gel according to differences in pI (isoelectric focusing)
Changes in pH can alter charge
Can disrupt secondary, tertiary, and quaternary structure (disrupt side chain interactions)
Can interfere with normal protein function
Zwitterion form
Zwitterion form = form in which overall charge on amino acid is 0
pI
Isoelectric point (pI) = pH at which overall charge on amino acid is 0
Duplications
(chromosomal mutations)
Duplications: involve the copying of DNA sequences that may remain in chromosome
Duplicated gene can evolve independently of original
Duplication can give rise to multiple subtypes of same kind of protein
Examples = different kinds of calcium channels, different subunits of hemoglobin
Major evolutionary mechanism
Transposons
(Chromosomal mutations)
- Transposons (transposable elements) = “jumping” sequences of DNA
- Can disrupt a gene or contribute to gene activation or silencing
Central Dogma
DNA replicates to make identical copies of itself
Genes coded in DNA are transcribed into RNA, another nucleic acid
RNA is translated to protein consisting of amino acids
Information flow: DNA makes RNA makes protein
A few exceptions to central dogma
In retrovirus, RNA reverse-transcribed to DNA with reverse transcriptase enzyme
Chromosome structure and chromatin
Eukaryotic chromosomes are linear
Only 1–3% of genome is contained in exons
Eukaryotic DNA is packaged as chromatin
Fundamental unit of chromatin is the nucleosome
DNA wraps around nucleosomes every ~150 base pairs
Nucleosomes are composed of (+) charged histone proteins
8 histones (4 pairs) per nucleosome
Chromosome structure and chromatin 2
histones have + charges on lysines and arginines (basic amino acids)
Positive charges interact with negative charges on phosphate groups of DNA
Also hydrogen bonds and non-polar interactions between histones and DNA
Histones can be modified by methylation, phosphorylation, acetylation, etc
Modification occurs on histone tails
Modification can involve addition of methyl group to lysine or arginine side chain
Chromosome structure and chromatin 3
CH3 attaches to amino group of side chain
Can involve addition of acetyl group to lysine side chain
Can involve addition of phosphate group to serine, threonine or tyrosine side chain
Chromatin modification can alter gene expression
Higher order packing of chromatin into loops fits DNA into nucleus
Euchromatin = loosely packed form of DNA, for regulatory regions as well as genes that are transcribed
Heterochromatin = tightly packed form of DNA (in Barr bodies, centromeres, and telomeres)
Euchromatin
Euchromatin = loosely packed form of DNA, for regulatory regions as well as genes that are transcribed
Heterochromatin 2
Heterochromatin = tightly packed form of DNA (in Barr bodies, centromeres, and telomeres)
Centromeres
Centromere is a key chromosome region
Essential for faithful chromosome segregation in mitosis and meiosis
Appears as a “pinched” region on condensed chromosome
Centromere is gene-poor and composed of repeat sequences
Centromeres = heterochromatin
During mitosis and meiosis, binds special proteins that form the kinetochore
Kinetochore is site of microtubule attachment
prokaryotic transcriptional control: operons
In prokaryotes, several genes typically transcribed together = operon
Genes in an operon generally operate in the same pathway
Example: Lactose operon contains several genes for lactose metabolism
Lac operon is transcribed only when lactose is present (inducible)
Operon is transcribed by default
Operator DNA sequence is binding site for lac repressor protein
Nearby promoter is blocked by bound lac repressor
No lactose → lac repressor binds operator and prevents transcription
Lactose present → lactose binds to lac repressor, causes dissociation from the operator
RNA polymerase can then bind promoter and transcribed operon
Glucose is preferred sugar, if present will repress lac operon
tryptophan operon
Example: Tryptophan is synthesized when it is not already present (repressible)
Operon is repressed by presence of tryptophan
When tryptophan is not present, repressor becomes inactive, genes in operon transcribed
Eukaryotic transcriptional control
Eukaryotic genes often controlled at the level of transcription
Some genes are always transcribed (constitutively expressed)
Other genes selectively transcribed
Default state of eukaryotic genes is generally OFF
Need additional control proteins to turn genes ON
promoters
Promoter is DNA sequence that dictates where RNA polymerase starts transcription
Conserved sequences in promoter are important for RNA polymerase binding
–35 TATA box
enhancers
Enhancers are DNA sequences that increase expression from a promoter
Can be distant from promoter (can be > 50 kb away)
Can be located upstream, downstream, or in introns
May also confer spatiotemporal or tissue-specific expression (e.g., brain)
Enhancers are binding sites for transcription factors
Combination of promoter and control elements controls expression pattern
transcription factors
Transcription factors are proteins that bind to control regions near genes
Can activate (increase) expression, or sometimes decrease gene expression
Activators help recruit RNA polymerase to gene
Silencers prevent RNA polymerase binding, alter chromatin
Two functional parts of TF proteins: DNA binding, transcriptional regulation
Transcription factors can be tissue-specific (e.g., brain, liver, heart)
Regulate transcription factors → regulate gene expression
Transcription factors work in combinations
Transcription factors often regulated by signal transduction or hormones
methylation and transcription
A methyl (–CH3) group can be added to bases in DNA
Methylation is generally associated with inactive genes (not expressed)
(Contrast with acetylation, which usually makes genes more likely to be expressed)
Post-transcriptional control
Alternative splicing
Translational control: keep mRNA around and translate when necessary
Control of protein activity via phosphorylation or zymogen activation
Subcellular localization of proteins can be regulated (e.g., move to nucleus)
Proteins can be degraded (e.g., cyclins in cell cycle)
Cancer
Cancer results from loss of control in cell proliferation
Two classes of “cancer genes”: proto-oncogenes and tumor-suppressors
Proto-oncogenes are genes that normally promote growth
A gain-of-function mutation in a proto-oncogene yields an overactive form (oncogene)
Oncogenes increase growth and proliferation
Tumor-suppressor genes normally restrict growth and proliferation
Loss-of-function mutation in a tumor-suppressor yields a less active or inactive form
Certain mutations may promote further mutations (e.g., defects in DNA repair)
a gain-of-function mutation
cancer
A gain-of-function mutation in a proto-oncogene yields an overactive form (oncogene)
Oncogenes increase growth and proliferation
loss-of-function mutation
cancer
Tumor-suppressor genes normally restrict growth and proliferation
Loss-of-function mutation in a tumor-suppressor yields a less active or inactive form
Certain mutations may promote further mutations (e.g., defects in DNA repair)
DNA Base pairing image
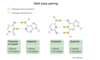
where does transcription occur?
IN THE NUCELUS for eu
In eukaryotes, transcription and translation take place in different cellular compartments: transcription takes place in the membrane-bounded nucleus, whereas translation takes place outside the nucleus in the cytoplasm.
In prokaryotes, the two processes are closely coupled
Base pairing for H bonds
Which base has the most hydrogen bond DONORS:
GUANINE
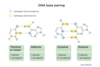
Hydrogen bond donors image
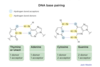
deoxythymine!
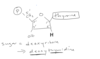
acetyl group added to Lysine
image of promoter and enhancer in dna
Question 25- QBank 11-25
Alpha helices display a measurable molecular dipole along their long axes. This property results from:
A.hydrogen bonding between spatially adjacent R groups on the exterior surface of the helix.
B.hydrogen bonding between backbone carbonyl oxygen atoms and protonated nitrogen atoms separated in sequence by several amino acid residues.
C.hydrogen bonding between backbone carbonyl oxygen atoms and protonated nitrogen atoms from amino acids that are directly adjacent in sequence.
D.hydrogen bonding between spatially adjacent R groups on the interior face of the helix.
Question 25
Alpha helices display a measurable molecular dipole along their long axes. This property results from:
A.
hydrogen bonding between spatially adjacent R groups on the exterior surface of the helix.
This answer choice is incorrect. R groups vary greatly in character, and many do not have the ability to form hydrogen bonds at all. Since all alpha helices display the property described, it most likely does not result from such a wildly variable factor.
B.
hydrogen bonding between backbone carbonyl oxygen atoms and protonated nitrogen atoms separated in sequence by several amino acid residues.
B is correct. In an alpha helix, all carbonyl groups are oriented in the same direction; this results in a slight dipole moment. While these groups are not consecutive in terms of the amino acid sequence, they are spatially adjacent. This allows them to hydrogen bond.
C.
hydrogen bonding between backbone carbonyl oxygen atoms and protonated nitrogen atoms from amino acids that are directly adjacent in sequence.
This answer choice is incorrect.
D.
hydrogen bonding between spatially adjacent R groups on the interior face of the helix.
This answer choice is incorrect. Amino acid side chains are directed outward, away from the long axis of the helix.
In the final product of this process is a free phosphate group, which reagent must be added to complete teh reaction and free the substrate from teh enzyme?
H20
must be added to hydrolyze the substrate from the enzyme;s active site. the water molecule will donate a proton to teh cysteine residue and a hydroxyl group to the phosphate ion. Note that water is often used to promote hydrolysis reactions, which break bonds.

H bond acceptors vs. donors
Remember:
Hydrogen bond donors are only those H atoms bound to an electronegative atom such as N or O.
Hydrogen bond acceptors are electronegative atoms with at least one lone pair of electrons.


