Cycle 10 - DNA Technologies Flashcards
(21 cards)
Describe the basic mechanism of propagation of action potentials in neurons
- Neurons are polarised: positive on the inside, negative on the outside
- Lets positive ions rush in to depolarise the membrane
- Creates the action potential
Describe how channelrhodopsin can control action potentials in neurons.
- Used in optogenetics
- Channelrhodopsin from chlamy was placed in a rodent, and shining blue light around it opens the channel, sending the action potential all with the shining of light
Describe some possible sources of genes (ie. channelrhodopsin) for use in making transgenic organisms.
- One source is the chlamy genome: cutting it into pieces with restriction endonucleases, which can be ligased to create recombinant DNA molecules
- mRNA could be harvested from chlamy (easy), then it is reversed into double stranded DNA through reverse transcriptase creating cDNA (no introns)
- Simply typing the sequence out into a gene synthesizer
State the role of restriction endonuclease enzymes in creating recombinant DNA
- Restriction enzymes cleave sugar-phosphate backbones and create sticky ends
- The restriction in the name of the enzymes refers to their normal role inside bacteria, in which the enzymes defend against viral attack by breaking down (restricting) the DNA molecules of infecting viruses
- The bacterium “hides” the restriction sites in its own DNA by attaching methyl groups to bases in those sites, thereby blocking the binding of its restriction enzyme.
- Restriction enzymes read 5’ to 3’, and are symmetrical (palindromic)
- If DNA cut by the same restriction enzymes are mixed together, the sticky ends will base pair and are sealed by DNA ligase creating recombinant DNA molecules in a plasmid vector
- These vectors are offered to bacterial cells (transformation: taking up DNA from the environment)
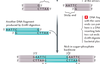
Describe the basic mechanisms for creation of cDNA
- Primer attaches to a sequence of mRNA
- Reverse transcriptase synthesizes a complementary strand of DNA
- The mRNA degrades and DNA polymerase synthesizes a strand of DNA complementary to the other strand
- Result is cDNA: a DNA molecule that is complementary to an mRNA molecule, synthesized by reverse transcriptase
Describe ways in which cDNA versions of genes would be different than genomic versions
cDNA would not contain introns or splice signals, but would contain a polyT tail (as well as the complementary polyA tail)
This is useful for cloning in bacteria, which cannot remove introns by themselves
Describe the role of antibiotic resistance genes on vectors in genetic engineering
When you offer DNA to bacteria, only a selection will take up the vector
All cells that take up the vector will become resistant to ampicillin (ampR gene), so ampicillin is used to kill the others
Describe the mechanisms used for introducing transgenes into microbes, plants, and animals
- Plasmid & cDNA methods as described above
- “Biolistic” gene guns can deliver transgenes to plant nuclei and organelles by shooting them
- Rhizobium radiobacter has a plasmid called Ti which goes right into the nucleus of plants (a bacteria that naturally shoots DNA into plants) –> we put DNA of our choice into the Ti plasmid and let the radiobacter work
- Some viruses like adenovirus deliver DNA into the nucleus
- Was used to deliver CFTR to CF patients but with only temporary effects
- Microinjection: inject DNA into the nuclei of the cells
Describe the role of reporter genes (eg. GFP) in genetic engineering
Explain GFP, LacZ, and luciferase
- Introduce reporter gene along with transgene of interest, if the organism shows reporter gene activity, it means the transgene was introduced
- GFP glows green under UV when present, so it can be used to see if a rat has taken up the desired gene entirely
- LacZ: a bacterial gene that can show its presence by creating blue pigment
- Luciferase: fireflies manufacture light using luciferase
Explain the strategies for expressing channelrhodopsin in specific neurons
- Put the gene and its tissue-specific regulator (promoter) into the organism
- Put channelrhodopsin under a neuron-specific promoter
- Use a tissue-specific virus – virus only inserts gene into neurons
Explain the DNA sequence “ChR2eYFP” / “optogenetic construct”

- ITR: inverted terminal repeat, important for incorporation of the DNA into the genome of the rat
- EF1a: promoter, strong
- ChR2: channelrhodopsin from chlamy, fused with
- eYFP: yellow fluoresces protein, reporter gene
- WPRE: extends the half life of mRNA, from a woodchuck virus
- hGH: human growth hormone polyA signal/clipping sequence
Describe the composition of expression vectors
- Gene of interested (cDNA) inserted into restriction sites
- Promoter, transcription terminator, SD box
- Antibiotic resistance (ampR gene) - selectable marker

Describe the basic mechanism of PCR (Draw)
- PCR takes one double strand of DNA and amplifies it
Steps
- Denatured into single strands
- Add in DNA primers (useful for specificity) which bind to the 3’ ends and moves toward 5’
- DNA polymerase extends the primer
- We use Taq polymerase which is useful because it functions at high temperatures which is necessary for PCR (several points of temperature increase to denature)

Say I have some DNA: 5’GTTAA3’ what would be the primter for this?
3’CAATT5’ or 5’TTAAC3’
Describe the usefulness of transgenic (recombinant) strategies in research as well as commercial protein production.
- Useful in basic research: investigate the role of a gene
- Useful in commercial products/industrial products
- Useful for medical treatments
How does bacterial photography work?
- Introduce genes that help produce light sensitive protein PCB in E. coli (originally blind)
- Introduce gene for kinase → phosphorylates ompC promoter → increase expression of lac Z → produces black pigment
- Presence of light → kinase undergoes conformational change and does not phosphorylate → no black pigment
State the branches of Canadian government responsible for regulating production and sale of genetically modified organisms
Health Canada
Agriculture and Agri-Food Canada
Environment Canada
Define GMO according to Canada
- “One derived from an organism that has had some of its heritable traits changed by”
- Traditional techniques of cross breeding
- Chemical or radiation mutagenesis
- Recombinant DNA techniques
- So… everything we eat
Describe the basic process of transgenic “Arctic Apples”
- Browning of apples occurs from the activity of polyphenol oxidase genes
- Artic apples have stopped browning by creating transgenic trees
- They use a Ti plasmid (moves genes into the nucleus of plants) that has the polyphenol oxidase gene
- Has the CMV promoter (cauliflower mosaic) which is very strong
- Results in crazy gene expression of the polyphenol oxidase gene
- If our cells get infected by a virus, we get a huge spike in viral mRNA
- Over evolutionary time, our cells have figured out that huge spikes in mRNA = virus!
- So, our cells shut down the gene that is extremely active
- Thus, the regulatory machinery shuts down the polyphenol oxidase gene when it is spiking due to CMV, and viola, no browning
- Has kanamycin resistance in order to identify the cells with the vector
Describe the basic process of transgenic “AquaBounty Salmon”
- Triploid, all female Atlantic salmon, first GM animal approved for human consumption in Canada
- Grow faster
- This is because it has a growth hormone gene under the control of an antifreeze gene from ocean pouts, which produce antifreeze constitutively (all the time)
- We take the constitutive promoter and hook it up to the normally seasonal growth hormone gene, and thus the fish grow to market size faster
Describe the creation of a knockout mouse
- Take cells out of a mouse embryo and grow them in culture, add in our gene of choice (ex., channelrhodopsin), one cell takes it out and becomes antibiotic resistant –> we grow the desired cell and kill the others
- We take these cells and inject them into a mouse embryo, the mother births them and they are called mosaics (made up of normal embryo and transgenic cells; only expressed in some regions), and some genes may be in the gametes
- Thus, when we cross two mosaics together, we get some babies that are homozygous for channelrhodopsin
- To identify those with channelrhodopsin, we insert a reporter gene


