Unit 3 - Cell Biology of Cancer Flashcards
function of CTLA-4
brake on T-cell activation
functions to regulate T-cell activation
cancer cells benefit from reduced T-cell activation
MAb vs CTLA-4 releases the brake, allowing enhanced T-cell killing of tumour cells

PD-I
required for T-cell activation
acting through a different mechanism PD-I also acts as a brake on tumour-directed cells
MAb vs PD-I also ‘releases the brake’, allowing enhanced T-cell killing of tumours

use of MAbs
- MAb vs CTLA-4
- MAb vs PD-I
treatment with MAb has led to dramatic clinical outcomes - remissions and cures of metastatic cancers
- releases brake ⇒ enhanced T-cell killing of tumour cells
- releases brake ⇒ enhanced T-cell killing of tumour cells
CAR T-cell therapy
Chimeric Antigen Receptor
T-cells (specialised WBCs) are isolated from a patient and a custom designed gene, that expresses a new cells surface molecule that recognises the tumour and activates the T cell to kill it, is introduced into cells
cells containing the gene are grown in culture to prepare an inoculum
CAR T-cells are infused back into patient
T-cells target cancer cells for killing
MOA of CAR T-cell therapy

what is cancer
a disease that originates at the cellular level but tumoue function as complex tissues that integrate multiple cellular functions and mechanisms to promote tumour survival and growth

what is needed to identify the cellular origin of tumours
histology
how do cellular properties change as cancer develops/progresses
acquisition of adaptive phenotypes through mutation and genome instability couples with recruitment and modification of non-cancer cells to form tumour microenvironments
⇒ for diagnosis and prognosis + understanding therapeutics, knowledge of the cellular basis of cancer is good pragmatic knowledge (personalised therapy)

6 Hallmarks of Cancer
- sustained proliferative signalling
- evading growth suppressors
- activating invasion and metastasis
- enabling replicative immortality
- inducing angiogenesis
- resisting cell death

metastasis
migration of tumour cells from primary tumour to secondary sites
responsible for 90% of cancer deaths
how do cells spread
via blood, lymph and through proximity
where might secondary tumours form
lung, bone, liver, brain
lymph nodes
what are secondary tumours
tumours of primary tissue irrespective of tumour site
e.g. breast cancer within liver
histochemistry can identify tumour type and aid design of treatment
invasion-matastasis cascade - 7 steps
- localised invasion
- intravasation (into circulation)
- transport
- arrest (in a secondary location)
- extravasation (out of circulation and into tissue - colonisation)
- proliferation
- colonisation
utilise mechanisms related to pathways of embryonic development and wound healing

malignancy
penetration of tumour c ells beyond basement membrane id definitive of malignancy
EMT
epithelial → mesenchymal transition
change in phenotype
properties of epithelial cells
polygonal morphology
network of cell-cell junctions
apical-basal polarisation
limited mobility/motility
mesenchymal cells - properties
migratory
variegated morphology/spindle shaped
loosely organised
present in connective tissue/stromal tissue e.g. fibroblasts

key components of EMT
expression of embryonic transcription factors e.g. Snail, Slug, Twist, Zeb 1/2
loss of e-cadherin function
loss of tight junctions
acquisition of motility through CT
protease secretion
growth factor receptor expression
EMT - change in markers
Epithelial cells express epithelial markers and do not express mesenchymal markers
Twist - down regulation of epithelial cell markers and upreg of mesenchymal markers
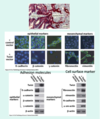
anchorage-dependent signalling
E-cadherin
functions as a cell adhesion molecule
Maintains epithelial cell phenotype by signalling cell-cell interactions via IC domain
loss leads to dysregulation of β-catenin, a transcription factor regulated by localisation in the cell

β-catenin
integrated into cadherin-actin adherens junctions complexes
a normal component of Wnt signaling pathway
upon loss of cell adhesion it translocates to nucleus to activate TCF/LEF family transcription factors - loss causes cell to move into a different phenotypic state
regulated by molecular association e.g. E-cadherin and APC and by inhibitors e.g. ICAT (inhibition of β-catenin and TCF4)
cytoplasmic levels are maintained through ubiquitin-dependent proteolysis via the β-catenin destruction complex
mutation/misexpression correlated with cancer progression

familial adenomatous polyposis
proliferation of polyps in colon
1 in 30,000
APC gene
function = regulation of β-catenin through the proteolytic pathway
tumour suppressor gene
autosomal dominant mutations
maintains epithelial cell phenotype in colonic crypts
integrates cellular architecture, motility with cell cycle regulation and gene expression
also functions in mitosis and loss contributes to CIN
(cells live for 4 days)




































