Unit 1 - Signal Transduction Flashcards
Molecular
activation of enzyme, generation/synthesis of metabolite and its degradation
cellular
number of mitochondria regulated in a cell may divide, split, fuse
tissue/organ
no of cells or cell types are tightly regulated
4 levels of regulation in living organism
molecular
cellular
tissue/organ
organism (neuro-endocrine)
main mechanims of integration of the levels of regulation
HPA axis

what is the target of regulation

definition of signal transduction
process linking the signal-activated receptor and the biological response
there is a common logic in the structure of signal transducers and in the mode of their function
how is regulation initiated and and how does it flow
initiated by the signal (information) reaching the cell or formed within the cell

EC signal molecule → receptor protein → IC signalling proteins → target proteins

what can a signal be, based on their origin
from environment - pin, temp, light, smell
from organism - hormones, cytokines, metabolites
from within the same cell - DNA damage, ROS
what can a signal be if chemical
- proteins - GH
- peptides - insulin, neuropeptides
- AAs, derivatives - thyroxine, dopamine, epinephrine
- lipids - PGs, platelet activating factor
- ions - Ca2+, Cl-
- nucleotides - adenosines with specialised receptors on their surface
- gases - nitric oxide (produced by endothelial cells – trigger smooth muscle relaxation and vasodilation)
types of signal transmission

what is the significance of ligand binding
specificity
amplification
co-ordination of response - the same receptor may be present on a number of cells
cell specific response - by regulating the number and function of receptors
example of disease that involves a cell-specific response
type 2 diabetes
autoinhibitory receptors are present on cells become inactivated and unresponsive
types of receptors
cell surface/transmembrane receptors
nuclear receptors
cytosolic receptors

receptors for what hormone are present on a wide variety of cells
cortisol
amplification system
Uniform response of cell
e.g. If a cell triggers lipid degradation, whole lipid synthesis across the cell has to shut down and lipid degradation must occur
Net 0 effect - what 1 process achieves, the other will undo

allosteric regulation

interconversion cycle
NOTE: enzyme specific - some are active phosphorylated, some are active dephosphorylated
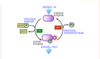
Akt/Protein Kinase P interconversion cycle - 1st mechanism
inactive Akt → Akt by 2 mechanisms:
FIRST
phosphorylated serine 473 interacts with the linker between the kinase and pH domains

2nd mechanism in Akt/protein kinase P interconversion cycle
phosphorylated serine 477 and threonine 479 result in the displacement of the pH domains

enzyme cascade

acceptors
all signal transduction pathways culminate to control of function of proteins (mostly enzymes, ion channels, transporters), which are involved directly in formation of biological response
2 categories of acceptors
control of protein amount
gene expression
protein degradation
protein stabilisation (tumour suppression gene P53)
control of existing proteins
without covalent modification of the protein (P) - Allosteric activator binds to an allosteric enzyme complex
with covalent modification of the protein - reversible and irreversible (cleavage of 3 forms of enzymes e.g. digestive enzymes involved in cleavage of trypsinogen to inter trypsin)
tumour suppression gene P53
regulated by protein stabilisation
mechanisms of hormonal regulation

4 types of membrane bound receptors
ion channel enclosing receptors
7 transmembrane domain receptors
1 hydrophobic domain receptors (R with enzyme activity, R with no enzyme activity)

ligands that activate membrane bound receptors
hydrophilic
signalling via ion channel-enclosing receptor example
nicotinic ACh receptor
structure of nicotinic ACh receptor
binding results in
pentamer (2 x α, β, γ, δ)
subunits surround a central pore
2 Ach binding sites from the pore
their binding is co-operative and leads to channel opening
channel is selective for divalent cations (+ve) - Na+ and Ca2+
hyperpolarisation with AP triggered

ION CHANNEL-ENCLOSING RECEPTORS
GABA, glycine, ACh, glutamate, serotonin
- receptors
- amplification systems
- acceptors
- biological response
- on the plasma membrane
- channel-transmitted ions
- membrane ion channels (e.g. VG Na+ channels)
- membrane depolarisation, hyperpolarisation, muscle contraction etc
glycine ion selectivity
Cl-
HCO3-
GABA ion selectivity
Cl-
HCO3-
ACh ion selectivity
Na+
K+
Ca2+
glutamate ion selectivity
Na+
K+
Ca2+
serotonin ion selectivity
Na+
K+
ION CHANNEL-ENCLOSING RECEPTORS
IP3, cGMP, cAMP, ATP
- receptors
- amplification system
- acceptors
- biological response
- on IC membranes
- channel-transmitted ions
- myosin, membrane ion channels
- muscle contraction, depolarisation, hyperpolarisation, activating of energy producing metabolic pathways also
IP3 ion selectivity
Ca2+
cGMP ion selectivity
Na+
K+
cAMP ion selectivity
Na+
K+
ATP ion selectivity
K+
overview of signalling system of ion channel-enclosing receptors

example of signalling via 7-transmembrane domain receptors
β adrenergic receptors
structure of 7 transmembrane domain receptors
amino terminal lies on EC side
carboxyl-terminal is in the cytosol
ligand sits in a pocket formed by transmembrane helices
upon R activation, the R binds a G protein (GPCRs)
3rd TM domain recognises the G protein

amplification system of G-protein activated adenylate cyclase

how does protein kinase A regulate the acceptor
PKA when activated by cAMP can penetrate the nuclear membrane
It can then phosphorylate CREB
When phosphorylated, it is ACTIVE and helps to initiate gene expression

acceptors of protein kinase A
- enzymes
- structural proteins - troponin, myosin light chain kinase
- ion channels - IP3-sensitive Ca2+ channel
- transcription factors (cAMP response element binding protein - CREB)
biological responses induced by adenylate cyclase system
in what structures is it present
Liver
adipocytes
muscle
adrenal cortex
T cell
Olfactory cells
crypt cells
SM cells
biological responses induced by adenylate cyclase system
LIVER
glycogenolysis
glyconeogenesis
glucolysis
gluconeogenesis
lipid synthesis
β oxidation
cholesterol synthesis
biological responses induced by the adenylate cyclase system
ADIPOCYTES
lipid breakdown
biological responses induced by the adenylate cyclase system
adrenal cortex
synthesis of many steroid hormones
ACTH regulated aldosterone secretion
biological responses induced by the adenylate cyclase system
OLFACTORY CELLS
sensitivity to odorants
biological responses induced by adenylate cyclase system
CRYPT CELLS
Cl- secretion
biological respones induced by adenylate cyclase system
SM CELLS
inhibition of contraction
biological responses induced by adenylate cyclase system
MUSCLE
glycogenolysis
biological responses induced by adenylate cyclase system
T CELL
proliferation
death
Amplification system - G protein activated phospholipase C
IP3 = ion channel

catalytic rxn of PLC
PLC can be activated via G proteins or by Tyr phosphorylation
membrane bound phosphatidyl 4, 5 bisphosphate is hydrolysed to membrane bound diacyl-glycerol and cytosolic ionositol 1, 4, 5-trisphosphate

activation of PLC?
via G proteins or by Tyr phosphorylation
amplification of G protein activated PLC

targets of calcium
calcium activates protein by directly binding or binding to a calcium-activated regulatory subunit
acceptors that are directly activated by Ca2+
tissue transglutaminase
protein kinase C
phospholipase A2
calpain
DNases
calcium-sensing regulatory subunits
functions
calmodulin regulated proteins
glycogen phosphorylase kinase, muscle contraction, calmodulin regulated protein kinase
AAs associated with PKC
Ser
Thr
targets of PKC
cell surface receptors - EGF, insulin, CD3
enzymes - raf1 kinase, GAP-p21 ras
ion channels - Na/H+ exchange
proteins in cell cycle control - DNA topoisomerase
nuclear factors - NFκB
proteins in cytoskeleton - MARCKS
PLC-induced biological responses
liver glycogenolysis (adrenaline α1, vasopressin)
thrombocyte aggregation (PAF, thromboxan)
thrombocyte serotonin secretion
mastocyte histamine secretion (IgE)
pancreas digestive system enzyme secretion (cholecystokinin)
insulin secretion
adrenal chromaffin cell adrenaline secretion
adrenal cortex aldosterone secretion (Ang II)
SM contraction (PGE2)
cell proliferation
differentiation
programmed cell death
signalling system via 7-transmembrane domain receptors

signalling through 1-hydrophobic domain receptors carrying enzymatic activity
- give an example

receptor tyrosine kinases (in their C terminus) - insulin receptor

receptor tyrosine kinase
how many present
how do the domains differ
what are the receptors for
58 in humans, in 20 sub-families
1 transmembrane region
variable EC ligand binding domain
IC tyrosine kinase domain
receptors for growth and survival factors - drive cell proliferation, cell survival, cell differentiation

GOF mutation in TK receptor
GOF ⇒ constitutively active in the absence of their ligand
they will drive constant cell proliferation, cell survival making these cells resistant to drugs
ligands of TK receptors induce
RTK dimerisation, leading to activation

what do the phosphorylated tyrosines act as
docking sites for signal transducing proteins

phosphotyrosine residues act as binding sites for
enzymes and adaptor proteins
what are adaptor (scaffold) proteins
proteins with platforms to bind multiple proteins
ability to bring protein complexes together
proteins with SH2 (Src homology domain 2) and PTB (phosphotyrosine binding) domains can dock on phosphorylated tyrosines of RTK
the type of adaptor protein determines the downstream signalling and the biological response
3 main signal transduction pathways induced by RTKs

what is Ras
what amplification cascade does it activate
a small GTPase, similar to G proteins
activates the MAPK amplification cascade
Ras activating enyme
Raf (Ser/thr kinase)
RAS signalling drives
cell division
Ras and cancer
Ras (HRAS, NRAS, KRAS)
one of the most frequently mutated oncogenes in human cancers

what is Ras unable to do
single subunit protein so not able to replace GDP with GTP
hence GEF protein helps with this - removes GDP and replaces it with GTP
Ras pathway
Grb2 (scaffold) binds to P-Y via its SH2 domain
Grb2 has 2 SH3 domains, which bind Sos
Sos = guanine nucleotide exchange factor that activates Ras
Ras activates Raf
Raf is a serine threonine kinase and activates mitogen activating protein kinases

how and what does Ras induce

3 main Ras proteins in the body
KRAS
NRAS
HRAS
Ras mutations and cancer
GOF - affects GTP binding
phosphatidyl ionositol 3 kinase pathway
- how is it activated
by RTKs
(PLC is seen both with 7 TM receptors but also RTKs)

what does PI3K phosphorylate
the membrane lipid phosphatidyl ionositol 2-phosphate

structure of PI3K
regulatory and catalytic subunit
both subunits have multiple protein binding motifs and recognition motifs (SH2, SH3)
1 of the most frequently mutated pathways in human cancers

phosphatidyl ionositol 3 kinase pathway

FoxO
blocks cell signalling
Forkhead transcription factor
regulates expression of Bim
GLUT4
glucose uptake into cells
activated by Akt
GSK3
glycogen synthase kinase
Akt inhibits GSK3
metabolic adaptation
leads to glycogen metabolism, cell cycle progression
mTOR
metabolic adaptation
protein synthesis for cell proliferation
(activated by Akt)
Bad, Bim
cell survival
Akt inhibits Bad, Bim, Bax (which are all pro-apoptotic)
what does activated Akt do
regulates receptors via phosphorylated serine and/or threonine residues on them
what are inhibitors of PI3K used to treat
chronic lymphocytic leukaemia
PI3K and Akt are
proto-oncogenes
(GOF ⇒ constitutively active - Drive proliferation, drug resistance)
signalling system of receptor tyrosine kinases

3 groups of signal transduction pathways
membrane bound receptors
cytosolic receptors
nuclear receptors
types of membrane bound receptors
ion channel-enclosing receptors
7-TM receptors (β adrenergic)
1 hydrophobic domain receptors (with or without enzyme activity)

requirement for IC receptors
ligands must be membrane permeable
examples of IC receptor ligands
steroid hormones
lipophilic vitamins
small molecules e.g. NO and H2O2

example of cytosolic receptor
what is its ligand
soluble guanylate cyclase receptor
receptor for NO

functions of NO
regulation of SM relaxation
platelet aggregation
neurotransmission

where does the synthesis of NO occur
in endothelial cells, neuronal cells and macrophages
enzymes that produce NO
what are their isoforms
nitric oxide synthases (NOS)
neuronal NOS
endothelial NOS
inducible NOS
** expression is not restricted to these tissues
how is the synthesis of NO controlled
by hormones, cytokines, bacterial endotoxins
VIA
- regulating the IC Ca2+ level (eNOS, nNOS)
- regulating the synthesis of NOS on gene level (iNOS)
soluble guanylate cylcase-induced signalling

what are nuclear receptors
transcription factors - intitiate gene transcription
nuclear receptors intitiating gene transcription

what are class I nuclear receptors
inactive receptor located in cytosol

what complex keeps the receptor inactive
HSP complex
what happens when the hormone binds to the nuclear receptor
dissociation of heat shock proteins
dimerisation
translocation to the nucleus
what family does the class I NR belong to
steroid receptor family
- progesterone - PR
- oestrogen - ER
- glucocorticoid - GR
- androgen - AR
- mineralocorticoid - MR

nature of changes in gene transcription mediated by steroid receptors
relatively slow in onset yet result in long-term changes in gene expression
where are class II nuclear receptors found
retained in the nucleus even when no ligand is bound
ligand binding to class II nuclear receptors causes
dissociation of corepressor protein
recruitment of coactivator protein
recruitment of additional proteins including RNA polymerase to the NR/DNA complex to transcribe DNA into mRNA resulting in biological response

examples of class II nuclear receptors
retinoic acid receptor
retinoid X receptor
thyroid hormone receptor
vit D receptor (VDR)
peroxisome proliferator-activated receptor (PPAR)


