Tour of MCB Block 1 Flashcards
(425 cards)
What is an enzyme?
Chemically active proteins (or RNAs)
What is the general name for the enzymes that copy a DNA strand during normal transcription?
RNA polymerase
How many chromosomes are carried by a normal diploid human cell?
46
What is the name for the barrier that separates the eukaryotic nucleus from the cytoplasm of the cell?
E. Nuclear envelope
Roughly how many protein-coding genes are found in humans?
20,000
Which of these processes is part of normal RNA maturation in eukaryotes, but not found in prokaryotes?
RNA splicing
HOw does a fully mature mRNA get into the cytoplasm of a cell?
It is exported through the nuclear pores.
The crossing-over observed betwwen chromosomes during human meiosis is better termed:
Homologous recombination
Approximately what percent of the human genome is repetitive DNA?
60%
Which of these processes is part of normal RNA maturation in eukaryotes, but not found in prokaryotes?
RNA capping
What is produced by meiosis I?
Two haploid cells containing bivalent chromosomes.
Are prokaryotic Genes “polycystronic”
NO
How much of the human genome is unique sequence?
40%
What is a SINE?
Short interspersed nuclear element
What percent of the human genome codes for functional RNAs?
1.6%
What of these is the best desription of the human mitochondrial genome?
Small 16,500 bp DNA circular chromosome
Why do we include coverage of bacterial plasmids in our course?
They are the main mechanism for antibiotic resistance in bacteria.
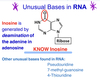
What is the correct name for a purine or pyrimidine base linked covalentlky to a pentose sugar?
Nucleoside
In adenosine monophosphate (AMP) where is the phosphate attached to the sugar?
at the 5’ position
What are the LINEs in the human genome?
Long interspersed nuclear elements.
What is a retrontransponson?
It is a genetic component that can copy and paste itself into different genomic sites-using RNA as an intermediate.
(using RNA as intermediate)
What is the size of the repeat unit in microsatellite DNA?
2-4 bp
What is the simplest name for this molecule?

Deoxyadenosine triphosphate
Which of these is a simpler way of writing pCpApTpGpGpC?
CATGGC