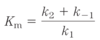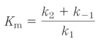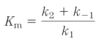Chapter 6: Enzymes Flashcards
(127 cards)
cofactor
- reusable non-protein molecules that doesn’t contain carbon (inorganic)
- Usually are metal ions such as iron, zinc, cobalt, and copper that loosely bind to an enzyme’s active site
- a substance that increases the rate of a chemical reaction
- can be considered “helper molecules” that assist in
- They must also be supplemented in the diet as most organisms do not naturally synthesize metal ions.
coenzyme
- an organic non-protein compound that binds with an enzyme to catalyze a reaction
- often broadly called cofactors, but they are chemically different
- it cannot function alone, but can be reused several times when paired with an enzyme
prosthetic groups
- organic vitamins, sugars, lipids, or inorganic metal ions
- unlike coenzymes or cofactors, these groups bind very tightly or covalently to an enzyme to aid in catalyzing reactions
- often used in cellular respiration and photosynthesis.
A complete, catalytically active enzyme together with its bound coenzyme and/or metal ions is called a _____. The protein part of such an enzyme is called the _____ or _____
- holoenzyme
- apoenzyme
- apoprotein
active site
- region of an enzyme that binds substrate molecules
- This is crucial for the enzyme’s catalytic activity.
substrate, S
The molecule that is bound in the active site and acted upon by the enzyme
The surface of the active site is lined with _____ _____ _____ with substituent groups that bind the substrate and catalyze its chemical transformation. Often, the active site _____ a substrate, sequestering it completely from solution.
- amino acid residues
- encloses
Catalysts do not affect reaction _____
equilibria
energy in biological systems is described in terms of
free energy, G.
ground state
- starting point for either the forward or the reverse reaction
- the contribution to the free energy of the system by an average molecule (S or P) under a given set of condition
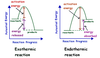
exothermic reactions
- the system loses heat as the surroundings warm up
- heat energy is being released from the system to the surroundings
- -ΔH = NEGATIVE
endothermic reactions
- the system gains heat as the surroundings cool down
- heat energy is being absorbed by the system from the surroundings
- ΔH = POSITIVE
exothermic reaction coodinate

- y axis = potential energy
- x axis = reaction pathway
- forward reaction: reactants on left, products on right
- reverse reaction: flip side reactants & products are on
- activated complex: is an intermediate compound w/higher energy than both reactants and products
- ΔH represents the difference between enthalpy of reactants and products
- ΔH = HPRODUCTS – HREACTANTS
- The step with the highest activation energy (ΔG‡) is the slowest step reaction
- The step with the lowest activation energy (ΔG‡) is the fastest step in the reaction
- The reaction cannot proceed faster than the rate of the slowest elementary step

endothermic reaction coodinate

- y axis = potential energy
- x axis = reaction pathway
- activated complex: is an intermediate compound w/higher energy than both reactants and products
- ΔH represents the difference between enthalpy of reactants and products
- ΔH = HPRODUCTS – HREACTANTS
- The step with the highest activation energy (ΔG‡) is the slowest step reaction
- The step with the lowest activation energy (ΔG‡) is the fastest step in the reaction
- The reaction cannot proceed faster than the rate of the slowest elementary step

reaction coordinate
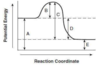
- A: potential energy of reactants
- B: ΔG‡: activation energy: energy needed to start reaction
- C: ΔG‡: activation energy: reverse reaction
- D: ΔH: energy of reaction
- E: potential energy of products

ΔGo, the standard free-energy change
- describes the free-energy changes for reactions
- a standard set of conditions
- temperature 298 K
- partial pressure of each gas 1 atm, or 101.3 kPa
- concentration of each solute 1 M
ΔG’o, the biochemical standard free-energy change
- used because biochemical systems commonly involve H+ concentrations far below 1 M
- it is the standard free-energy change at pH 7.0
reaction coordinate diagram

- S = Substrate, P = Product
- free energy of the system is plotted against the progress of the reaction S → P
- description of the energy changes during the reaction
- horizontal axis (reaction coordinate) reflects the progressive chemical changes (e.g., bond breakage or formation) as S is converted to P
- activation energies, ΔG‡, for the S → P and P → S reactions are indicated
- ΔG’o
- standard free-energy change in the direction S → P
- exergonic: it’s negative, the free energy of the ground state of P is lower than that of S
- at equilibrium there is more P than S (the equilibrium favors P)
transition state
- The rate of a reaction is dependent on an energy barrier between reactants and products
- energy required for alignment of reacting groups, formation of transient unstable charges, bond rearrangements, and other transformations required for the reaction to proceed in either direction
- depicted by the energy “hill” in reaction coodinate
- molecules must overcome this barrier with a higher energy level
- the top of the energy hill represents a moment where things could go either way, decay to either substrate or product is equally likely
- not a chemical species with any significant stability and should not be confused with a reaction intermediate
activation energy, ΔG‡
- difference between the energy levels of the ground state and the transition state
- rate of a reaction reflects ΔG‡
- a higher ΔG‡ corresponds to a slower reaction
- Reaction rates can be increased by raising the temperature and/or pressure, thereby increasing the number of molecules with sufficient energy to overcome the energy barrier
- ΔG‡ can be lowered by adding a catalyst
reaction intermediates
- A reaction intermediate is any species on the reaction pathway that has a finite chemical lifetime (longer than a molecular vibration)
- occupy valleys in the reaction coordinate diagram
rate-limiting step
- the step (or steps) with the highest activation energy determines the overall rate
- it’s the highest-energy point in the reaction coordiante diagram
- can vary with reaction conditions, and for many enzymes several steps may have similar activation energies, which means they are all partially rate-limiting
- Reaction equilibria is linked to the _____ _____-_____ for the reaction
- reaction rates are linked to the _____ _____
- standard free-energy change ΔG’º
- activation energy ΔG‡