1 - Intro To DNA Flashcards
What is the structure of DNA?
- Right handed double helix
- Antiparallel complementary strands held together by hydrogen bonds between complementary base pairs
- 2-deoxyribose
- Phosphodiester backbone
- Polar due to 5’ 3’ ends
- Major/minor groove

What is chromatin?
DNA wrapped around histones (octomer of proteins) , which can then condense to form chromosomes
What is the structure of euchromatin and heterochromatin?
Euchromatin: Light. Beads on a string, when DNA being replicated. Less dense as acetylated.
Heterochromatin: Dark, solenoids packed to form a 30nm fibre. Dense as methylated. Gene not expressed like this

What is the name of DNA and a histone together?

Where do proteins for DNA replication bind to DNA?
Major groove as easily accessible
What is the process of forming a chromosome?
- DNA wraps around histones to form nucleosomes and beads on a string
- Nucleosomes wrap around one another and form solenoids
- Solenoids join together to become fibres
- Fibres zigzag over one another to become highly condensed loops
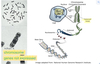
What is the genome?
- Entired DNA sequence
- 24 chromosomes, 2 sex 22 auto
What are nucleosides?
- Sugar and base, no phosphate
- Adenosine, Guanosine, Uridine, Deoxythymidine, Cytidine
What are the two classifications of nucleotide bases?
- Purines (2 C ring): A,G
- Pyrimidines (1 C ring): T,C,U
How would you find a gene on a chromosome if you knew the exact position?
LOOK AT THE BANDING PATTERNS
Why can C only bind to G?
- 3 hydrogen bonds
- All bonds of nearly equal distance
Why is DNA antiparallel?
Ensure the bases are close together and in the middle so they can form hydrogen bonds
What is the nomenclature of DNA?
- Left to right 5’ to 3’
- Complementary is 3’ to 5’
What is the cell cycle and where are the checkpoints?

What is the equation for DNA replication?
(dNMP)n + dNTP –> PPi + (dNMP)n+1
What does DNA polymerase do?
- Reads in 3’ to 5’ direction
- Elongates in 5’ to 3’ direction
- Catalyses phosphodiester bond using energy from PPi hydrolysis
- Can proof read

How is DNA replication started?
- Primase (RNA Polymerase) makes a primer which binds to the start of DNA wanting to be replicated
- DNA polymerase binds to primer to start elongation
What is the process of DNA replication?
1. Initiation

- Topoisomerase and DNA helicase unwind the double helix to form replication forks with two template strands (one leading one lagging)
- Primase produces short primers that bind to the 3’ ends of the DNA
2. Elongation
- DNA polymerase binds to primers and reads in 3’ to 5’ direction, adding complementary base pairs to the primer in a 5’ to 3’ direction
- On the lagging strand (5’ TO 3’ orientation) there are lots of primers so okazaki fragments are formed and replication is discontinuous
3. Termination
- Replication forks meet
- Primers removed and replaced with appropriate bases
- DNA strand proofread
- DNA ligase joins okazaki fragments
- Semi conservative replication
If DNA polymerase works at 100 nucleotides a second, and a DNA molecule is 6000 nucleotides long, how long will replication take?
- Impossible to know as there are so many replication forks running in different directions, not just one DNA polymerase
How do prokaryotes and eukaryotes differ?
Prokaryotes have:
- Smaller ribosomes
- Nucleoid (no nucleus)
- Plasmid
- Slime capsule
- Flagella
- Cell wall of muerin
- No membrane bound organelles
Why are nucleosides used for analogue drugs instead of nucleotides?
Nucleotides are charged so would be difficult to get them to cross the membrane


