Week 9.16: Environmental effects Flashcards
(17 cards)
Lifestyle Effects
Aside from food and toxins (i.e diet, cosmetics, recreational drugs), there are many other lifestyle factors known to affect health. For example;
<!--[if !supportLists]-->
· <!--[endif]-->Type and quantity of exercise
<!--[if !supportLists]-->
· <!--[endif]-->Sleep patterns
<!--[if !supportLists]-->
· <!--[endif]-->Stress
<!--[if !supportLists]-->
· <!--[endif]-->Posture
Even specific physical activity can be important. Think about it – someone may have a genetic predisposition to hip problems, but many such people will only have a single hip replacement. Why?
2. ** <!--[endif]-->Finding associations**
Environmental effects are hard to find
Model systems typically used to investigate the effect of specific factors, e.g toxicology testing of cosmetic compounds on animals or cell lines. Very limited and artificial, only shows generic changes – not different genomes. Does not show long terms effects interaction with other factos, etc.
Studied on human populations are essential -
Due to the way that humans have evolved, there are often confounding factors
<!--[if !supportLists]-->
· <!--[endif]-->People with similar genomes often live in the same area
<!--[if !supportLists]-->
· <!--[endif]-->People with similar genomes often have similar diets for cultural reasons, or because of genetic reasons (e.g lactose intolerance)
<!--[if !supportLists]-->
· <!--[endif]-->People with similar genomes often have similar behavioural habits, again due to culture or location.
<!--[if !supportLists]-->
· <!--[endif]-->Strong correlations between environmental factors and medical interventions e.g high sugar intake correlated with number of mercury fillings; people with allergies likely to take antihistamines
It’s therefore hard to separate genomic characteristics from environmental factors
What does make a good experimental design?
What does make a good experimental design?
<!--[if !supportLists]-->
· <!--[endif]-->Only change one variable at a time
<!--[if !supportLists]-->
· <!--[endif]-->No point measuring variables that are known to be well correlated
<!--[if !supportLists]-->
· <!--[endif]-->Use a control set to test the null hypothesis
<!--[if !supportLists]-->
· <!--[endif]-->Use replicates to determine variance
<!--[if !supportLists]-->
· <!--[endif]-->Confirm finding on spate cohort
Not east to design an experiment because, for ethical reasons, human subjects cannot be subjected to deleterious environmental factors. You can’t feed people obvious toxins. Indeed, anything shows to significantly increase mortality is off limits – e.g increasing vitamin E intake
Even if you can perform a population-wide study capable of finding a GxE relationship, for an individual you are unlikely to have sufficient environmental data to modify his/her disease risk.
Case study: Type 2 Diabetes T2D
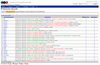
Patel et al. used a GWAS-type approach to produce the results below, suggesting serveral environmental factors that have strong associate with T2D:

These people who have had their data collected about this
This is one way of going about this study trying to relate GxE
Crucially, the effect sizes of the environmental factors are comparable to genomic loci found using GWAS.
Case study: Type 2 Diabetes T2D
How did they do this?
Data from US centre for disease control NHANES programme. Freely available data capturing huge amounts of information from a bunch of individuals via questionnaires, examination and laboratory analysis.
From this data, 266 environmental factors were considered in the T2D study. To avoid self-reproting bias, these were all from analytical assays, not questionnaires.
Survey-weighted logistic regression model corrected for known confounding factors; age, gender, BMI, socioeconomic status
One cohort used to build a model, other cohorts used to test validity.
CDC website where you can access this type of information
Logistic regression is a problematic classification technique, essentially mapping one or more variables to a particular binary (i.e two state) output (e.g case/control)

Case study: Type 2 Diabetes T2D
Permutation of cases and controls was used to estimate false discovery rate;
Sensible results?
Did they get any useful results from this? Benefit of beta-carotenes correoborated previous epidemiological studies outside the US, BUT, interstingly beta-carotenes not shown to be effective in treating T2D in clinical trials. Why?
Confirmation of negative affect of polychlorinated biphenyls PCBs, previously reported in non-US cohorts.
Gamma-tocopherol, a form of vitamin E, has a negative effect. More evidence of potential toxicity of vitamin E.
Heptachlor epoxide (a pesticide0 is a newly found factor.
Case study: Type 2 Diabetes T2D
**Limitations **
Expensive: Acquisition of the NHANES data from each volunteer is estimated at approximately $40,000 = far more expensive than genotype or even genome sequencing.
Associate NOT causation – T2D could be causing increased accumulation of certain chemicals, not vice versa.
No GxE: No genomic information used in this studies. This EWAS study is separate from GWAS studies. Genetic information now being collected by NHANES – this is the next step!
3. ** <!--[endif]-->Rare variants**
By the very nature of their rarity, the effect of rare variants cannot be revealed by the population-based studies like GWAS and EWAS
To determine possible GxE effects for such variants, we can use databases of relationships between genes and, for example, toxins.
Comparative toxicogenomics database
CTD integrates data from curated scientific literature to describe chemical interactions with genes and proteins, and associations between diseases and chemicals, and diseases and genes/proteins
If a novel variant is found in an individual’s gen, possible GxE interactions can be inferred by querying a database like CTD.
This is a starting point to predicting what effect the individual’s variant may have (be it positive or negative).

4. Your personal environment: the microbiome
** Metagenomics & the human microbiome**
To fully understand how a human being functions, we need to consider our own personal environment, not just external factors.
We couldn’t survive without help from microorganisms that we carry: bacteria, fungi and archaea. Up to 1,000 individual species of bacteria believed to live in the human gut alone.
Ten times as many bacterial cells as human cells, in the body, accounting for up to 3% of body mass.
Bound to have extensive interactions with the human genome.
G x M x E (gene x microbe x environment) interactions
Skin Microbiome
Images showing you different locations on the body –pie charts showing abundance of different bacteria in different locations on the skin
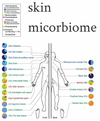
Difficult to characterise. Trying to really understand what is the microbiome environment – next generation sequencing techniques to characterise environment around the exterior of human beings
You have bacteria in all sorts of locations – including on your eyes
Ten times as many bacterial cells as human cells in the body, accounting for up to 3% of body mass…
Array-based techniques and NGS allow us to characterise the microbiome better than ever before.
Disruption of the microbiome already shown to increase the risk of developing cancer, type 1 diabetes and Crohn’s disease.
In mice, strong associations have been found between the genome and microbiome
Looking at relationships of any kind between genome variation and microbes – GWAS type way
In mice bacteria living in symbiosis with mice – this is showing the mouse genome and black stripes are individual SNPs
Figure shows mouse genome. Black lines indicate SNPs, Regions of the genome showing significant association with presence of bacteria are coloured according to the colour of the bacterium in the phylogenetic tree in the centre
From PubMed 20937875

Relationship between microbiome and environment less well understood, but there are some very obvious relationships:
<!--[if !supportLists]-->
· <!--[endif]-->Smoking – known to affect oral microbiome. Also shown to increase risk of Crohn’s disease.
<!--[if !supportLists]-->
· <!--[endif]-->Personal hygiene – could you be too clean?
<!--[if !supportLists]-->
· <!--[endif]-->Taking antibiotics.
Some big efforts underway to learn more about this, for example Human Microbe Project
Personal genetic risk modifiers
A big motivation for discovering GxE interactions is to identify actions that can be take to modify (reduce) disease risk!
Disease with a post-test probability of > 10% shown. Size of disease name indicates probability for the individual – black arrow indicates that one disease increases probability of another. Grey arrows indicate relationship between behavioural factors and disease…
“Hmmm, better do more exercise and stop smoking. Not really a mind blowing insight”
Information about how to modify behaviour so you don’t have a particular disease

The future?
Lots of hype about BIG DATA
Many sources of environmental information ready for the taking:
·Location data from your mobile phone
·Location data can be linked to weather, pollution data to build up a picture of exposure to heat, cold, UV, pollution.
·Location data can be mined to work out how often you visit the gym, go for a run, cycle to work etc.
·Diet habits from you payment cards/ loyalty cards
·Do you buy plenty of fruit, alcohol, cakes?
·Lots of eating out? Regular at Stepney friend chicken
This data is without doubt noisy, but still far better than anything that was possible before.

All the data is available
The future is already here

Ginger.io is a teach start up founded to do exactly this. Diabetes pilot project already underway with Sanofi
Gadgets like Jawbone Up and Nike FuelBand track activity sleep patterns, etc.
No large coordinate population-wide studies involved such data yet, but some significantly sized studies underway.



