Protein targeting and export Flashcards
(161 cards)
An average protein is 10nm across, what is the volume of the protein?
41 nm3 in volume
An average eukaryotic cell is 50 µm in diameter and has a volume of what?
6.6x1013 nm3
An overview of protein targeting and export

Whats the fluorescent protein used to visualise subcellular localisation?
What organism was it discovered in?
GFP
Discovered in Aequorea victoria (Nobel prize winning discovery)
What structure does GFP have and when it’s mutated in different ways what can be produced?
GFP has a beta-barrel structure
Different colours can be obtained by mutating the Beta-barrel structure
e.g. mutating into YFP, BFP, CFP
This is because the wavelengths can be manipulated whilst moving through the structure and therefore produce different colours

How else can different colours be obtained from GFP?
By using other organisms such as DsRed
What can GFP fuse with?
Your proteins of interest (chimera- A chimera is essentially a single organism that’s made up of cells from two or more “individuals”)
Whats a disadvantage of GFP?
Chimeric protein may not fold correctly/ act normally due to large size of FP
The use of Immunofluorescence (IF) has the same kind of procedure as what?
Immunoblotting (detection of a protein on an antibody)
What are the disadvantages of Immunofluorescence?
- Cells have to be fixed as permeabilised to allow antibody into them i.e. dead
- antibodies may give false signals via non-specific binding

What are some other useful fluorescent dyes and what do they detect?
- DAPI (4’,6-diamidino-2-phenylindole) and Hoechst stain bind to the minor groove of DNA and emit blue light when exposed to UV light
- Phalloidin toxin from the death cap mushroom (Amanita phalloides)- binds to filamentous Actin (F-actin) and can be conjugated to different fluorophores

What are some of the reasons that cells migrate?
What does it require a rapid change in?
What needs to be newly synthesised?
- Wound healing, movement of WBCs to infection sites, metastasis
- Requires rapid changes in the cytoskeleton
- The leading edge of the cell has lots of actin microfilaments
- These assemble/disassemble to push membrane forward
- New synthesis of actin protein is also required to help in the migration process

Whats a FISH?
A probe with a fluorophore to show where the RNAs are
Whats are UTRs in eukaroytic mRNA?
They are Untranslated regions which are involved in regulation

sequences in the 3’ UTR may be involved in what?
localisation
e.g. the beta-actin mRNA contains a “zip code” which has an essential ACACCC sequence
What does the Zip-code in eukaryotic mRNA form?
A secondary structure via base-pairing within the 3’ UTR
Tell me what the Zip code biding protein (ZBP1) contains?
And what do these bind to?
- Contains 2x RRMs (RNA recognition motif) which has basic regions (positive charge) and interacts with negative charge on RNA
- Also, 4x KH domains (K-homology, first found in hnRNPK) which bind to single-stranded RNA and DNA

What does ZBP1 bind to?
What is this complex then transported through?
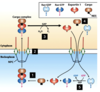
ZBP1 binds to nascent Beta-actin mRNA in nucleus as part of a large mRNP complex
These mRNPs are actively transported through the nuclear pore complex (NPC) with the aid of Ran GTPase
How is beta-actin mRNA transported to the leading edge of a migrating cell?
Where are many proteins made before being moved to where they need to be?
What do they therefore require?
Many proteins are made in the cytoplasm and then are moved to where they need to be
They require organelle-specific targeting sequences
How can organellle-specific targeting sequences be identified?
Some can be identified from primary sequence (NLS or NES)
Some are more structural
- amphiphilic helix (mitochondria)
- Signal patch (lysosome)
With organelle-specific targeting when entering the nucleus, what is required?
Tell me about them
A targeting sequence known as nuclear localisation signal (NLS) which is NOT removed following transport
There are two kinds of NLS which may be anywhere in the protein
- Basic
- Non-basic
What are the two kinds of NLS?
- Basic
- Non-basic
Whats a Basic NLS and give an example
Basic
e.g. SV40 large T antigen
- rich in lysine (K) and arginine (R)
- The exact sequence is not important instead it is the cluster of basic amino acids
- can be bipartite (split into two parts)