integuments Flashcards
what are dental integuments?
examples?
‘substances that form on the surface of teeth ‘
- Coating of developmental origin
- Cuticle, reduced enamel epithelium, coronal epithelium
- Acquired coatings
- Pellicle, plaque, calculus
what is the pellicle?
what are its components?
Protein layer on the enamel surface formed very quickly
components
- proteins from
- saliva, GCF, serum
- bacteria and their metabolic products
- connective tissue metabolites
how is the pellicle layer developed?
- First layer forms in <5 seconds
- Strong interactions
- Low molecular weight proteins laid down first
- Pellicle layer matures over days
- Mature pellicle is 10µm thick with surface and subsurface layers
what proteins are found in the dental pellicle?
- Lactoferrin
- IgA
- Salivary amylase
- Histatin
- Statherin
- Cystatin
- Acidic proline rich phosphoprotein
- Albumin
- MG1
which forces lead to pellicle formation on enamel??
adhesive forces
- Ionic interactions
- Van der Waals forces
- Hydrogen bonds
- Hydrophobic interactions
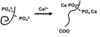
how are salivary proteins attached to the enamel surface by ionic bonds?
Electrostatic attraction between ionised groups:
- negative groups COO- or PO43- on protein to Ca2+, HPO42- on Hydroxyapatite (HAP) surface
- Strong binding

what does ionic bonding between the salivary proteins and enamel surface cause?
what are the consequences of this?
- Interaction between hydroxyapatite and calcium ions leads to change in conformation of the phosphoprotein
- Exposes hydrophobic regions
- Often binding regions for bacterial receptors
what are the functions of the pellicle?
- Acid protection
- Enamel protection
- Assists in re-calcification of enamel defects
- Provides a continuous layer of protein at the enamel-gingival interface
- Provides a surface for the adsorption of bacteria
- bacterial colonisation of the pellicle
what are the stages of plaque formation?
- Initial transport of bacteria to tooth surface
- Reversible adsorption of the bacteria onto pellicle surface
- Less reversible attachment of bacteria & matrix synthesis by bacteria
- Sugar matrix with cross linked sugars
- Attachment becomes less reversible
- Growth of attached organisms forming a bacterial community containing many “distinct” species.
describe the transport of bacteria to the tooth surface
Transport of micro-organisms to pellicle-coated tooth via passive means
- Local variations in the chemical composition of pellicle can influence the pattern of microbial deposition

describe the initial interactions between the micro-organisms and the enamel surface
reversible interactions
- formation of long-range physico-chemical interactions between micro-organisms and pellicle proteins
- electrostatic (ionic and H-bonds)
- van der Waals forces (induced dipole formation)
- micro-organisms are usually negatively charged
- acidic proteins usually in pellicle
- interactions - formation of calcium bridging
describe the short range interactions between bacteria and pellicle
Less reversible attachment of bacteria to tooth surface
- short range interaction between
- adhesins on microbial surface
- proteins in pellicle
- usually specific
- usually very strong
- irreversible
what is required for bacteria to bind to the pellicle proteins?
- For plaque formation to proceed it is necessary that not all bacteria aggregate in saliva before they reach the tooth surface
- Conformational changes of proteins on adsorbed surfaces allows bacteria to bind
- Pellicle protein binds to enamel surface and exposes the binding site for bacteria
what bacterial binding proteins are there?
- bacterial adhesins
- serin-rich repear proteins (Srr)
- antigen I/II polypeptides (Ag I/II)
- Pili/fibrillar adhesion proteins
how do bacterial adhesins cause bacterial binding?
what is their structure
- Specialised ‘glue’ proteins
- General concept of substrate recognition by bacterial adhesins
- Interactions with pellicle proteins

how do serine-rich repear proteins (Srr) cause bacterial adhesion?
- contains large amount of serine residues
- has recognition site for sialic acid in the mucin MG2
- leads to adsorption of streptococci to enamel surface
- drop in pH -> alters adhesive properties

how do antigen I/II polypeptides (Ag I/II) cause bacterial adhesion?
- gp-340 = immunity scavenger receptor & pattern recognition molecule
- bound to enamel
- recognised by bacterial antigen I/II polypeptides
- causes adhesion to enamel surface
- has different variants
- can increase adhesion and caries risk
- isopeptide bond stabilises the structure of the protein
- peptide was used to vaccinate caries

which binding protein was used for vaccinations for caries?
was this successful?
antigen i/II polypeptides (Ag I/II)
- unsuccessful
- Multiple adhesion receptors on the streptococci
- Targeting one receptor is not sufficient to prevent disease
how do Pili/Fibrillar adhesion proteins cause bacterial adhesion to the enamel surface?
- Composed of 3 subunits
- Found on Gram + and – bacteria
- 3 distinct protein subunits
- up to 3 micrometre long (very large)
- can bind to extracellular matrix and cell surfaces
- some bind to salivary pellicle proteins, i.e. proline rich protein
- Also type 2 FimA binds Gal-GalNAc-containing surfaces
- host cells
- oral microbiota
- Required for complex biofilm formation
describe the F. nucleatum and Strep. Mutants complex
- Adhesin required for strep.mutants interaction
- adhesin RadD recognises strep.mutants
- Complex is formed between strep.mutants and F.nucleatum
- foundation is laid for the building of the cities of slime….
- Interaction between RadD and strep.mutants can be interrupted with arginine, a positively charged amino acid.
- P.gingivalis can coaggregate to this complex
- Occurs through FomA protein
- Implications for periodontal disease
- Initiates inflammation of the gingiva



