Retroviruses, reverse transcription, retrotransposons Flashcards
Describe the contributions of Peyton Rous.
Peyton Rous was the first to show that a virus (RSV1), isolated from chickens, was capable of causing solid tumors of chickens in 1911. He won the Nobel prize for this work in 1966 and died the same year (1970) that the RSV oncogene was identified by Duesberg and Vogt.
Describe the contributions of David Baltimore and Howard Temin.
Baltimore and Temin simultaneously published results on the discovery of reverse transcriptase activity (RNA-dependent DNA polymerase). Baltimore independently isolated reverse transcriptase from RNA tumor viruses MLV and RSV. Temin discovered reverse transcriptase in RSV. Baltimore and Temin shared the Nobel prize with Renato Dulbecco in 1975.
What are the general structures of a retroviral genome?
R Region: A short (18-250nt) sequence that forms a direct repeat at the both ends of the genome, which is
therefore ‘terminally redundant’.
U5: A unique, non-coding region of 75-250 nt which is the first part of the genome to be reverse
transcribed, forming the 3’ end of the provirus genome.
Primer Binding Site (PBS): 18nt complementary to the 3’ end of the specific tRNA primer used by the virus to
begin reverse transcription.
Leader: A relatively long (90-500nt) non-translated region downstream of the transcription start site and
therefore present at the 5’ end of all virus mRNAs.
Polypurine Tract: A short (~10) run of A/G residues responsible for initiating (+)strand synthesis during
reverse transcription.
U3: A unique non-coding region of 200-1,200nt which forms the 5’ end of the provirus after reverse
transcription; contains the promoter elements responsible for transcription of the provirus.
What type of genome do retroviruses have?
Retroviruses have a positive-strand ssRNA genome. Each virion carries 2 copies of its genome.
What is an advantage of using lentiviral vectors in the lab?
Lentiviruses are able to infect differentiated, non-dividing cells.
What are the conserved retrovirus protein-coding domains?
- gag encodes virion proteins that form the matrix, capsid, and nucleoproteins
- pol encodes the reverse transcriptase
- env encodes the surface and transmembrane components of the viral envelope
What does the gag domain encode?
The gag domain encodes viral assembly proteins.
What does the pol domain encode?
The pol domain encodes reverse transcriptase and integrase.
What does the env domain encode?
What does the pro domain encode?
The pro domain encodes the viral protease. It is a smaller domain than the other protein-coding domains of the retrovirus.
Describe the steps of retroviral reverse transcription.
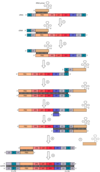
What are the subunits of reverse transcriptase?
Reverse transcriptase is made up of two subunits: p51 and p66 that have molecular weights of 51 and 66 kDa respectively. p51 is derived from p66 by proteolysis.

Describe the functional domains of reverse transcriptase.
The p66 subunit largely contacts nucleic acid substrates. This subunit contains regions called fingers, palm, thumb, connection, and the RNaseH domain. In the active site, the fingers close against the palm to sandwich the RNA template. Mg2+ ions are located in the active site and are required for enzyme activity (although Mn2+ can substitute with about 1/2 efficiency).

What are retrotransposons?
Retrotransposons are regions of a host genome that show homology to retrovirus genomes but lack functional env genes. They are capable of replicating, but do not form infectious particles.
What are some classifications of retrotransposons?
- Ty retrotransposons are found in Yeast and are shown to replicate through an RNA intermediate.
- copia is found in Drosophila and was transferred from melanogaster to willistoni, possibly through a parasite.
- gypsy is found in Drosophila and was recently reclassified as a retrovirus after infectious particles formed that allowed gypsy to infect female Drosophila oocytes.

