Lecture 12 Flashcards
1) What is minimal media? Provide an example of a Bacterial strain that is able to utilize this type of media. 2) What is complex media?
1) A growth media with a single sugar and any necessary minerals, also known as “chemically defined media”. E. Coli can grow in culture on glucose as the sole C/E source when inorganic phosphate, ammonia, sulfate, some salt, and a bit of iron are provided.
2) A growth medium that contains many preformed components ready to transport and use directly in anabolism and energy production, i.e., a variety of amino acids, sugars, peptides, vitamins, nucleosides. etc.
UTR?
RBS?
1) Untranslated region
2) Ribosome binding site

1) What are transcriptional activator proteins?
2) What are transcriptional repressor proteins?
3) How do they work?
1) Many promoters also use a regulatory protein, however, some of these promoters lack a recognizable -35 sequence. Activator proteins help RNA poly core enzyme recognize and bind to the promoter.
2) Repressor proteins stop gene expression by preventing RNA poly core enzyme from recognizing and binding to a promoter or by blocking its movement down the DNA.
3) Activator and repressor proteins work in pairs (dimers) binding at two sites on the DNA. Two copies of the same protein work together. Each monomer has two domains. One domain is the recognition helix, which binds to DNA, and the other is the stabilizing helix, which holds the two dimers together. The dimers bind at inverted repeats in the DNA sequence.

How do transcriptional activators and repressors bind to the DNA? What does each helix do?
- Activator and repressor proteins that operate as dimers have a helix-turn-helix (H-T-H) structure.
- One helix (DNA binding domain or recognition helix) of each monomer associates with the DNA, within the major groove.
- The other helix (stabilizing helix) allows protein-protein contacts that hold the monomer together as a dimer.

1) How do activator and repressor proteins control transcription?
2) What is induction?
3) What is repression?
4) How do they work?
1) The do so by binding with small molecules (effectors); the effectors are signal molecules, either from the environment or made by the cell. The binding of an effector changes the conformation of the protein, so that it can then bind to another monomer and bind to, or be released from, a promoter. The binding of the regulatory protein/effector complex to a promoter induces or represses transcription of the genes under the control of that promoter.
2) Induce = increase transcription, often strongly.
3) Repress = decrease transcription, often strongly.
4) By helping RNA poly to find the promoter and bind to it (induction), or blocking the promoter so that RNA poly cannot find or bind to it or cannot move down the DNA (repression)
Gene regulation in bacteria is a decision making process. How do bacteria decide whether or not to turn a gene on or off?
- A process by a which a bacterium considers the current situation.
- What nutrients are available to it, what is the pH, temperature, salinity, oxygen level in the environment, what its current needs are for survival and reproduction.
- Based on what is available to the cell it then “decides”, at the molecular level, whether to turn on or turn off the expression of various sets of genes whose protein products are beneficial or not needed under the currect conditions.
- Bacteria are always and continutally assessing their current situation and deciding, at a very fast and fine scale, what genes to express and how fast and how long, and what genes to not express.
What is the moltose operon (E. Coli)? What is the inducer? What is the rational for this positive regulation?
- An example of positive transcriptional regulation (induction).
- Maltose = glucose-glucose dimer
- malE, malF, and malG code for proteins involved in binding of maltose in the peroplasm and transport of this sugar across the cytoplasmic membrane.
- inducer = maltose or maltotriose
- Turn on transcription of genes when the compound it works on is present.

1) What is negative transcriptional regulation? What is the rationale behind it?
2) Physiological analysis based on picture. What happens when:
a) do not add arginine?
b) add arginine at time = t?
c) add arginine at time = 0?
d) add alanine at time = t?
3) Measure of time: cells, total protein (why?) and arginine biosynthesis enzymes?
4) Repression - a typical way of regulating the production of enzymes involved in biosynthesis, why?

1) The rationale is to turn off synthesis of enzymes for making arginine if arginine is available from the environment.
2) assay for arginine biosynthesis enzymes for cell grown in minimal medium with glucose as the sole carbon and energy source.
a) continued biosynthesis
b) Arginine biosynthesis enzymes flatline
c) Flatline
d) Continued increase because alanine has nothing to do with this.
3) Looking to see if addition of arg affects all proteins or just biosynthesis enzymes.
4) Shut off biosynthesis when you do not need it.
Genetic analysis (genetic mechanism of repression)
1) What does the argCBH operon code for?
2) What is the structure of the operon?
1) Encodes enzymes [ArgA, ArgB, ArgH] involved in synthesis of arginine from biochemical precursors.
2) A promoter
An operator: Binding site within or adjacent to a promoter region where a repressor protein binds
RNA poly (argCBH had a σ70 type of promoter
Repressor protein and corepressor.
Lactose Operon Background information
- Concept: Jacques Monod and Francois Jacob
- lactose: a disaccharide (glucose and galactose); milk is a good source of lactose.
- E. Coli is a common inhabitant of the intestine of humans and other mammals.
- lactose is a good carbon/energy source for E. coli, but not as good a just glucose alone, because the cell has to modify lactose to split it into glucose and galactose (split by galactosidase), and these sugars are then used as carbon and energy sources.
1) What does the lac operon consist of?
2) What does lacZ, lacY, and lacA code for?
1) promoter, operon, lacZ, lacY, lacA and a terminator
2) lacZ: codes for ß-galactosidase, lacY: codes for lactose permease (transports lactose across the cytoplasmic membrane into the cell. lacA: codes for transacetylase (necessary for utilization of lactose)
Transcription of the E. coli lac operon is under dual control. negative and positive.

ß-galactosidase synthesis in E. Coli
1) Is the enzyme regulated or is it produced constitutively?
2) What would be the pattern if the enzyme was produced constitutively?

1) Regulated, constitutive means synthesis of the enzyme is not regulated; the amount of enzyme increases in direct proportion to the number of cells and the amount of total protein as the culture grows.
2) The same as cell number and total protein.
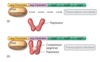
What is the mechanism behind the regulation of the lac operon?
- Negative control by a repressor protein.
- Repressor - LacI protein, expression of the lac operon is off (repressed) in the absence of an inducer.
- Inducer: allolactose, an isomer of lactose, formed from the lactose by a secondary activity of ß-galactosidase (derepresses transcription of the lac operon).
Even when expression of operon is derepressed transcription of the lacZYA genes is not strong, and cells make very little ß-galactosidase. Why?
- 35 sequence TTTACA (consensus = TTGACA) one difference from the consensus.
- 10 sequence TATGTT (consensus = TATAAT) two differences from the consensus.
- These differences suggest that the lac operon promoter is weak. Experimentally it has been found that binding of the RNA poly to the lac promoter is relatively weak. Weak binding of RNA poly at a promoter means weak transcription of downstream genes.

What is better for the cell to grow on? Glucose or Lactose?
In E. coli, glucose is a better C/E source than lactose or any other sugar; E. coli grows faster on glucose than lactose or any other sugar. So it would be wasteful for the cells to produce enzymes for the transport and catabolism of oter sugars when glucose is available.
Diauxic growth (two-growth)
1) Why is there a lag in growth after the cells have consumed the glucose?
2) Why do cells grow at a slower rate on lactose than they did on glucose?
3) What causes the level of ß-galactosidase to rise?

- The first part of the graph is an example of glucose repression - the presence of glucose somehow blocks the synthesis of ß-galactosidas.
- During the first part of the experiment, glucose is being used as the C/E source.
- When glucose has been consumed, the cells shift to using lactose.
1) Stop growing and begin synthesis of ß-galactosidas.
2)
3) The presences of lactose.
1) What does glucose repression indicate?
- Glucose repression (catabolite repression) is an indication that another form of control, a positive control, is present in this gene regulatory system.
- The positively acting control element is a transcriptional activator protein, called cyclic AMP receptor protein (CRP). Works together with an effector, cyclic AMP (cAMP). Positive Control!
- 3’:5’-cAMP is produced from ATP by the enzyme adenylate cyclase (coded for by the cyaA gene).

1) What is CRP the product of?
2) What does cAMP do to CRP?
3) What does cAMP-CRP complex do?
1) Product of the crp gene.
2) cAMP binds to CRP, changing its conformation so that it can bind, as a dimer, to the lactose operon promoter.
3) Helps RNA poly find and bind to the lactose promoter. The cAMP-CRP binding site is upstream of the lac operon -35 sequence. The consensus CRP binding site is: TGTGA-N6-TCACT notice that it is an imperfect inverted repeat.
What does glucose control?
- Glucose controls the level if cAMP in the cell by regulating the activity of adenylate cyclase, and by influencing excreation of cAMP.
- In the presence of glucose, adenylate cyclase activity is low and cells excrete cAMP.
- SO: when glucose is present, cAMP levels in the cell are low and the cAMP-CRP complex does not form. When glucose is not present, cAMP levels in the cell are high, and the cAMP-CRP complex forms.
Key point: transcription of the lac operon is controlled by (1) repression-derepression of the lacI protein and allolactose (negative control) and by (2) the level of cAMP-CRP (positive control).
What has to happen for RNA poly to transcribe the lac operon at a high rate?
1) the operator must not be occupied by lacI, so that movement movement of the polymerase down the DNA is not blocked, negative control factor is absent.
2) The CRP binding site must be occupied by cAMP-CRP, positive control factor present, to help RNA poly easily find and bind to the promoter.
- Need both at the same time for high transcription.
What is the logic of dual control? What conditions have to be met?
- The lac operon is transcribed at a high rate (the cell makes lots of ß-galactosidas and lactose permease) only when both of two conditions are met:
1) when lactose is available
2) when a better C/E source than lactose, such as glucose is not available. - It is important to understand the logic of the dual control of the lac operon of E. coli.