Lecture 11 Flashcards
(16 cards)
1) Central dogma and informational macromolecules
2) What are the three types of RNA?
3) What is translated mRNA, rRNA, or tRNA?
1) DNA is transcribed into RNA.
2) mRNA, tRNA, and rRNA.
3) mRNA is translated into protein. tRNA and rRNA is not translated.
1) Bacterial Genome?
2) What is genotype?
3) What is phenotype?
1) - All the genetic information of an organism chromosome and plasmids.
- E. Coli, Gram-negative bacterium. Genome size - 4.64 (megabases) = 4,640,000 bases (nucleotides) . Approximately 4300 genes
2) the gene by gene description of an organism’s genetic information.
3) the observable characteristics of an organism.
Transcription in Bacteria? What is the holoenzyme?
RNA polymerase (core enzyme of synthesis RNA) + sigma factor, σ (recongnizes site for RNA synthesis to begin) = holoenzyme

Each gene or set of genes has what?
1) a promoter, to guide the beginning of transcription.
- RNA poly unwinds the DNA at the promoter, the sigma factor falls off, and the core RNA poly beings to make a single strand of RNA that is complementary to the template strand.
2) the gene or genes that are transcribed
- The DNA is rewound after RNA poly passes.
3) a terminator
Termination of transcription
- RNA poly releases from the template DNA at specific termination sequences.
- Termination sequences often are a GC-rich region with an inverted repeat and a central non-repeating section.
- This arrangement leads to formation of a “stem-loop” structure just upstream of a run of U (uridine, RNA for T, thymine)
- RNA poly pauses when the stem-loop forms and falls off the template DNA at the run of uridines. This is called an intrinsic (or Rho protein-independent) terminator.

Termination of transcription - 2 (Rho protein-dependent terminator)
- Another kind of transcription terminator.
- Rho protein moves down the new forming RNA and when RNA poly pauses at the termination site Rho protein causes the RNA and RNA poly to come off the template DNA.
1) What is a Gene?
2) What is a Operon?
1) DNA coding for a single polypeptide (or for a tRNA or an rRNA).
2) a set of contigous genes under the control of a promoter.

What is a cistron?
- A cistron is a gene.
- Polycistronic RNA: the single transcript (mRNA) of the multiple genes of an operon.

Bacterial gene expression
Is it careful and smart?
What happens when environmental circumstances change?
What of the benefit of these changes?
- Gene expression is careful and smart.
- When environmental circumstances change, bacteria detect these changes and modify their production of the kinds and amounts of proteins to those that are useful or necessary under the new environmental condition.
- The benefit is competition (getting nutrients, energy etc. efficiently)
Gene regulation, what happens?
- Regulation of the kinds and amounts of different proteins made occurs at several levels in the cell.
- Different genes are transcribed at different rates, and those rates vary depending on conditions and the cell’s needs.
- mRNA is degraded; different mRNAs are degraded at different rates.
- Different mRNAs are translated at different rates.
- Activity of enzymes is regulated, as in feedback inhibition.
- In bacteria, regulation of transcription is the most important.
Regulation of the transcription of genes.
- In Bacteria, the transcription of genes (gene expression) is tightly and carefully regulated so that:
→ the right proteins are made.
→ in the right amounts
→ at the right time (i.e. when the cell needs them and can use them)
- Lower levels of those proteins are made when the cell does not need them in high amounts.
- Bacteria are exquisitely sensitive and responsive to changes in their environments,
- To survive (compete) they must utilize energy and nutrients from the environment as efficiently as possible to make more cells.
- Carefully regulating gene transcription saves energy and nutrients - the cell makes only proteins it needs when it needs them.
How does RNA poly find the start of the gene, open the double helix, and copy the template strand into RNA?
- By recongnizing the promoter - the region preceding a gene or operon, where RNA poly binds to start transcription.

Conventional numbering system for promoters of Bacteria
- RNA poly holoenzyme recongnizes specific sequences before (upstream) of a gene or operon, the -35 sequence and the -10 sequence (Pribnow box). These sequences are made up of two sets of six nucleotides separated by approximately 16-18 nucleotides. They define a promotor in Bacteria.
What are housekeeping genes?
- Code for proteins that are essential for normal cellular metabolism and reproduction.
- Use a sigma-70 promoter
- 35 sequence consensus is TTGACA centered ca. 35 bp upstream of transcription start (+1)
- Sigma factor: functions in promoter recongnition, -35 region.
- 10 (Pribnow Box) centered ca. 10 bp upstream of transcription start. RNA poly binding consensus TATAAT, favors unwinding of DNA.
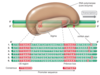
Variation around the consensus? What does this mean for a promoter to be strong or weak?
- Some variation around -35 and -10 sequences and for the space between these sequences.
- Also, for sigma-70 promoters, the closer the -35 and -10 sequences are to the consenses, the stronger the promoter
- The more the sequences differ from the consenses, the weaker the promoter.
- The weaker the promoter the more difficult it is for RNA poly to bind and begin transcription.
E. Coli
- 7 different sigma factors, for promoters of different kinds of genes - “housekeeping” genes (code for proteins involved in normal growth), sigma-70; other sigma factors help RNA poly recognize promoters for genes involved in nitrogen assimilation, stationary phase, heat shock response, flagella synthesis, etc.


