Lec 23 - Identifying Virulence Factors Flashcards
(21 cards)
Give the 3 ways we can identify virulence factors from pathogens
- IMMUNOLOGICAL; raise antibodies against immunogenic proteins that MAY have been produced in response to VF
- BIOCHEMICAL; need to know something about the virulence factor beforehand eg structure, isolates the antigen/VF and aims to determine function/activity
- GENETIC; does not require prior knowledge, most commonly used, invovles genetic approaches such as mutagenisis, complementation. Requires suitable animal model to test in
Describe the 3 methods of inducing RANDOM mutations
- irradiation
- chemical
- transposons - can insert randomly throughout the genome
Give an example of a genetic engineering method that can induce mutations in SPECIFIC locations around the genome and explain it in a little more detail
transposon mutagenesis
- eg Gram-ve can use Tn5 insertions
- mobile genetic elements
- Tns have transposase gene and antibiotic R marker
What are some advantages and disadvantages of using Tns to cause mutations
+ves
- introduces a single mutation
- non leaky - produces null mutants/knockouts
- easily selected using Antibiotic resistance marker
-ves
- polar effects on downstream genes
- does not insert into vital/essential genes (however ok because most VFs are not essential genes)
- may show bias to certain locations ad insert > frequently there
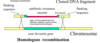
Outline the procedure used to find genes for cell invasion in Salmonella
- suicide plasmid (containing Tn) introduced into Salmonella. it cannot repicate in Salmonella
- Tn jumps into Salmonella chromosome and Salmonella grown on antibiotic
- colonies that grow contain Tns that have inserted into various locations throughout the genome
- each colony grown up and infected host cell
- look for colonies that cannot invade host cell
- amplify and clone the gene that has had the Tn insertion by 1) complementation OR 2) using antibiotic resistance as selectable marker to clone gene
What is the function of the suicide plasmid in Tn mutagenesis?
suicide plasmid can be propagated in E. coli but cannot replicate in the target organism therefore for Tn to replicate need to jump into the other chromosome
Give the OVERALL strategy of using genome sequences to identify VFs
- homologies to VF/gene sequences already known to find an unknown VF
- ampification using PCR and cloning of gene into vector and inactivation by insertion of antibiotic marker
- allelic exchange mutagenesis
- look for lack of VF / change in WT (compare WT and mutant)
Give the process of using antibiotic resistance casettes to identify VFs in organisms. draw a diagram to explain this
- gene for VF identified (potentially) in bacterium X
- amplification of this gene by PCR and cloning of this gene, along with some flanking regions, into vector (eg plasmid of E. coli)
- restriction enzymes remove part of this VF gene and replace with antibiotic R marker
- homologous recombination occurs when this plasmid transformed back into bacterium X and the WT VF gene replaced w/ antibiotic R marker
- mutant and WT phenotype can then be compared in suitable animal model
What are the 3 considerations taken into account when choosing an animal model ?
Route of infection needs to be the same as in humans.
Similar symptoms need to exist between the 2.
Type of animal model used
What happens when a pathogen cannot infect via the same route as an animal model?
compromised animal can be used
- gnotobiotic - lack of resident flora
- infants - immature immune system
- immunocompromised - NUDE (no T cells), SCID (no B/T cells), transgenic (no IFNy therefore no macrophage activation)
alternative route of infection
- intraperitaneal - injection straight into the body cavity however can’t asses invasion/colonisation
Give an example of when a different strain of bacteria causes a similar disease as in humans.
Whooping Cough
- Bordetella pertussis causes similar symptoms to Whooping cough (in humans) in primates however ethical issues surround the use of testing in primates
- a similar strain can be used to infect dogs/pigs (Kennel cough) in which similar symptoms shown - similar pathogenic mechanisms?
Why is it that pathogens do not always cause similar symptoms in the model?
- human specific
- exhibit different symptoms in animal model
Why are rice so popular in being tested on? (3 reasons)
- cheap and easy to maintain
- require limited growing space
- short generation time
What type of things / values can we obtain when testing animal models?
- LD/ID50
- bacteremia (no. bacteria in blood)
- histopathological changes - inflamation
- effects of mutants
What is the main requirement of these tests in order for the results to be statistically significant?
large no. animal test models
What are the advantages and disadvantages of using cell lines- tissue cultures to test early steps in infection?
+ves;
- single cell type
- easy to maintain and cheap to grow
- avoid animal suffering
- grown under reproducible conditions
-ves;
- often derived from tumour cells (immortalised, lack of cell traits and genetic changes)
- cell shape/polarity changes
- production of the culture medium (bile)???
Draw a diagram and include the processes and steps required to complete an adherene assay and invasion assay on a tissue culture

What is pictured here?

fluorescently labelled bacteria that adhere to the cell surface
How is a toxicity assay carried out?
- purified toxin
- infection of tissue culture
- looking @ LD50/ morphological changes
What are organ cultures and why are they sometimes considered better than using tissue cultures?
- tissue explants that contain a variety of cell types
- not just a single layer of cells
- not tumour derived (WT genetics) and contain normal cell shape/morphology


