introduction to epigenetics - lecture notes - julia Flashcards
(32 cards)
what is the definition of epigenetics?
- 1939: “the interactions of genes with their environment that bring the phenotype into being”
- 1987: terminology to define situations in which changes in DNA methylation result in changes in gene activity
- today: the study of heritable changes in genome function that occur without alterations to the DNA sequence
what are the three categories of epigenetic changes?
- DNA methylation
- histone modifications
- miRNA
what is DNA methylation? what mediates it? what nucleotides does it affect most often?
- enzyme-mediated chemical modification that adds methyl (CH3) groups at selected sites on the DNA double helix
- mediated by DNA methyl transferases
- in >99% of instances affects cytosine base (C) when it is followed by a guanine (G) = a CpG island
what is a CpG island?
- region most often methylated
- dinucleotides concentrated in specfic areas of DNA (300-3000 bp long)
- C followed by a G
- located in promoter region and first exon of genes
- methylation of CpG islands serves as a switch that can silence the downstream gene
- these islands are predominantly unmethylated in the healthy state!
what is the effect of DNA methylation? (ie how does it affect structure and protein interactions?)
- methyl groups protrude from the cytosine nucleotide into the major groove of the DNA
- displace transcription factors that normally bind to the DNA
- attract methyl-binding proteins which in turn are associated with gene silencing and chromatin compaction
what are miRNAs?
- small non-coding RNA
- 19-23 nt long
- single-stranded
- highly conserved
- complementary binding to their mRNA target
describe the steps in the biogenesis of miRNA
- in nucleus - transcription of large primary (pri) miRNA by RNA polymerase II or III
- these pri miRNAs are 100-1000 nt long and contain at least one miRNA stem loop
- may contain up to 6 (pre) miRNAs - RNAse III enzyme Drosha and microprossor complex unity DGCr8 cleave the pri miRNA to Pre-miRNA
- Pre-miRNA range from 70-90 nt in length and have hairpin structure - energy-dependent Exportin-5 transports them to the cell cytoplasm
- RNAse III enzyme Dicer crops the hairpin-shaped pre-miRNA to make a double stranded structure consisnt of the miRNA and its complement
- mature miRNA strand is incorporated into a miRNA associated RNA-induced silencing complex (miRISC)
- in this formation miRISC interacts with its target mRNA
- A) if the mRNA and the miRISC have perfect base pairing homology, mRNA will be cleaved and degraded
B) if the binding is imperfect (more common) there’s translational repression
what are the roles of epigenetics in the normal behavior of the cell? (5)
- x-chromosome inactivation
- silencing of parasitic DNA sequences
- correct organization of chromatin in active and inactive states
- tissue specific methylation
- genetic imprinting
how do miRNAs differ between normal tissue and neoplastic tissue?
- in normal tissue
- many miRNA are tissue-specific or tissue-enriched
- each tissue has a characteristic expression profile
- related tissues have similar profiles
- in neoplastic tissue
- miRNAs are differentially expressed
- selective miRNAs are up- or down-regulated in ALL tumor types
- other miRNAs are affected only in specific tumor types
- the site of origin of the tumor will have a characteristic miRNA profile
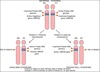
what is the difference between angleman syndrome and prader-willi syndrome? (in terms of symptoms and in terms of cause)
- in normal individuals, we get an active angleman gene from our mother, and the prader-willi gene is methylated to be inactive on the maternal chromsome, while the prader willi gene is active on the paternal genome and the angelman gene is methylated to be inactive (on chromosome 15q)
- summary of this:
- angelman gene is only active on the maternal chromosome
- prader-willi gene is only active on the paternal chromosome
- prader willi:
- mental retardation, short stature, hypotonia, hyperphagia, obesity, small hands and feet and hypogonadism
- have deletion on the paternal chromosome, so have an active angelman gene but no prader willi gene since the maternal copy of this gene is methylated
- angelman syndrome:
- mental retardation, ataxic gait seizures, inappropriate laughter, “happy puppets”
- have deletion on the maternal chromosome, so have an active prader willi gene but no angleman gene since the paternal verion of this gene is methylated
what is the genetic cause of fragile x syndrome?
- trinucleotide (CGG) repeat disorder
- x-linked
- repeats occur in the 5’ UTR of the familial mental retardation-1 (FMR1) gene
- phenotype related to the number of repeats
- normal is 6-31
- premutation 55-200
- full mutation - >200
- results in loss of function of the FMRP, which is a regulatory protein inportant for binding mRNAs, particularly in neurodevelopment
- hypermethylation results in silencing of this protein and abnormal development
*
what are the symptoms/clinical presentation of fragile x syndrome?
- 2nd most common genetic cause of mental retardation after down syndrome
- broad forehead
- long narrow face
- prominent ears
- hyper-extendible finger joints
- macro-orchidism (90% of boys by age 14)
- moderate or severe intellectual disability
- autism spectrum disorder
- epilepsy
- mitral valve prolapse
what are the epigenetic features of cancer cells? (3)
- hypermethylation of CpG islands of tumor suppressor genes
- global hypomethylation
- persistence of m5C residues => generation of spontaneous m5C to T mutations
what tumor suppressor genes does epigenetics seem to affect?
- inactivates
- BRCA1 in breast cancer
- Rb in retinoblastoma
- VHL in RCC
- p16 in solid tumors
- p15 in acute leukemia
what is MGMT? how does methylation affect it?
- DNA repair protein that removes mutagenic and cytotoxic adducts from O6 methyl guanine in DNA
- hypermethylation leads to gene silencing
- lots of tumors have high expression of MGMT
- patients with epigenetically silenced MGMT accumulate more mutations
- but epigenteicinactivation of MGMT enhances sensitivity to alkylating agents and is an indicator of better prognosis
- MGMT promoter methylation seen in 30-45% of high grade brain tumors
- basically - it’s confusing - not clear whether MGMT is good or bad…but if it’s methylated, we know we can treat it with alkylating agents
what is the consequence of methylation at the O6 position of guanine?
- important step in formation of mutations in cancer
- leads to conversion of G:C pairs to A:T => mutations in genes such as p53
what are MMRs? what do they do? how does methylation affect them? how does this impact the development of cancer?
- mismatch repair proteins
- correct any base mismatch errors that may occur during DNA replication
- about 15% of colorectal cancers have inactivated MMR DNA due to germline mutaitons or hypermethylation of the promoter
how is methylation involved in hereditary non-polyposis colorectal cancer?
- germline mutations inactivate a single allele
- somatic inactivation of the remaining normal allele by epigenetic events occurs (hypermethylation)
- => microsatellite instability and the development of tumors of the colorectum or endometrium at a young age
how is methylation invovled in the development of colorectal cancer?
- methylation of both alleles of the promoter or MLH1 occurs as a somatic event early in tumorigenesis
what is the general epidemiological information on renal cell carcinoma? (percentage of kidney cancers, all cancers? lethality? histological subtypes? differential diagnosis?)
- 85% of kidney cancer
- 3% of all adult cancers
- most lethal type of genitourinary cancers
- three main histological subtypes:
- clear cell (75%)
- papillary (10%)
- chromophobe (5%)
- beinign oncocytoma is an improtant differential diagnosis, especially for chromophobe carcinomas
how do the different types of renal cell carcinoma differentiated histologically?
- main idea here: it’s hard! biopsy samples are small and they all look similar - use molecular markers
- that said:
- clear cell has clear cytoplasm due to intracytoplasmic glycogen and lipids
- papillary has papillary architecture with foamy macropahges (bubbly cells)
- chromophobe has lots of cells that look really similar
- the oncocytoma is much pinker than the chromophobe and has less empty space
- (see following cards for actual images)
what would a clear cell carcinoma look like histologically?
- clear cytoplasm due to intracytoplasmic glycogen and lipids

what would a papillary carcinoma look like histologically?
- papillary architecture with foamy macrophages (the bubbly cells)

what would chromophobe carcinoma look like histologically?




