DNA replication Flashcards
DNA is replicated in semi-conservative replication, what is meant by semi-conservative replication?
- dsDNA separates into two single strands
- Each serves as a template for the formation of a new complementary strand
- so newly synthesised DNA consist of one old strand and one new strand, hence semi-conservative

What are the forces that hold the two complimentary strands together stably?
- H-bonds can form between complimentary bases
- A & T form 2 H-bonds
- C & G form 3 H-bonds
- H-bonds: relatively weak individually, but stable collectively
- DNA molecule has vast number of bases and H-bonds => stablity of dsDNA
Why is DNA’s structure in terms of its H-bonds ideal for replication?
- H-bonds weak => require relatively little energy to open them individually
Where does replication inititate?
- replication origins
- eukaryotes: multiple origins of replication
- bacteria: one origin of replication
What is replication origin (ori)?
- a special sequence in DNA that attracts initiator proteins
- A-T rich, because origin has to be easy to open
bacteria only has one ori, what mechanism do they use to speed of replication process?
stack replication: new replication bubbles start to form on newly synthesised DNA before the first replication bubble has even reached termination
Why do eukaryotes need multiple origins of replication?
- large genome
- replication of eukaryotic genomes is also slowed down by the complicated, compact packaging of DNA
- DNA cannot be replicated quick enough if there is only one ori
What does initator protein do?
- breaks H-bonds, opens DNA
- recruits replication machine (a cluster of proteins reponsible for DNA replication)
What are replication fork and replication bubble
- replication fork: the opening of two strands of DNA undergoing replication
- replication bubble: the bubble of unwound DNA formed between 2 replication fork
- multiple replication bubbles opens in DNA during replication

After initiation, elongation of DNA replication follows, carried out by replication machine. What are the proteins that make up the replication machine?
- DNA polymerase (DNAP)
- Primase
- Helicase
- Sliding clamp protein
- single strand binding protein
- topoisomerase
What is the function of DNAP
- synthesie new DNA
- has 2 active sites
- 5’ to 3’ polymerase => DNA synthesis = adds nucleotides to 3’ end
- 3’ to 5’ exonuclease => proofreading = removes misparied nucleotide
- note that DNAP can only synthesise DNA from 5’ to 3’ i.e. it can only add new nucleotides to 3’ end of polyneuclotide chain
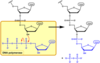
How does DNAP add new nucleotides to 3’ end of DNA?
- nucleotides enter reaction as free nucleosides triphosphates
-
phosphoanhydride bond hydrolysed
- releasing energy for condensation reation to form phosphodiester bond
- releasing two pyrophosphate (PPi)
- pyrophosphate then hyrolysed to inorganic phosphate
Difference between nucleotide and nucleoside?
- nucleoside: base + sugar
- nucleotide: base + sugar + phosphate group(s)
- nucleoside triphophate: base + sugar + 3 phosphate groups
Why can the DNAP only work in 5’ to 3’ direction
- Becasue DNAP cannot polymerise in 3’ to 5’ and at the same time have 5’ to 3’ proofreading activity:
- If DNA is synthesised in 3’ to 5’, it would be hydrolysis of the phosphate groups in the strand that provides the energy for formation of phosphodiester bond
- Removal of mismatched base would result in a 5’ OH group that has no phosphate groups attached, and so cannot provide energy to continue polymerisation

DNAP can only work in 5’ to 3’, but one strand of dsDNA is 5’ to 3’, which means its complimentary strand must be 3’ to 5’ (because dsDNA consists of antiparallel DNA molecules), how is the 3’ to 5’ strand synthesised?
- the 3’ to 5’ strand (lagging strand) is synthesied in 5’ to 3’ fragments called okazaki fragments
- okazaki fragments are later stitched together to from 3’ to 5’ daughter strand

What is primase?
- RNA polymerase
- adds primers (short sequence of RNA about 10 bp) that is base paired to the template strand
- Primers are later removed to be replaced by DNA

Why do primers need to added to template strand?
because DNAP cannot bind to ssDNA, need a double stranded end to start synthsising DNA
- dsDNA and ssDNA are chemically distinct situations
lagging strand has many primers cotinuously added to template, why?
- new primers are continually needed as DNA synthesis is discontinuous in lagging strand
- As movement of fork exposes new stretch of unpaired bases, a new RNA primer is made at intervals along lagging strand
How many primers does leading strand need?
one, at the origin of replication
How are primers removed and replaced by DNA?
- enzyme RNAse-H removes primers using a 5’ to 3’ exonuclease activity
- specific type of DNAP called repair DNAP replaces the gap with DNA
- DNA ligase then join the fragments together using ATP or NADH to form a continuous new strand
What is helicase?
- enzyme that unravels DNA further after initiator protein’s initial opening of DNA
- uses ATP
What is topoisomerase?
protein that prevents supercoiling of DNA
What is sliding clamp
-
keeps DNA polymerase firmly attached to the DNA template
- On lagging strand: releases DNAP from the DNA each time an Okazaki fragment is completed
- The clamp protein forms a ring around the DNA helix and binds DNAP allowing it to slide along a template strand as it synthesises new DNA
- increases speed of DNA synthesis
what are single strand binding proteins
- proteins that bind single-stranded DNA exposed by the helicase
- prevents it from reforming base pairs => prevents tangling






