DNA, Genetics, Evolution Flashcards
Differences between DNA and RNA

DNA structure, Crick and Watson, Rosalind Franklin
- DNA molecular structure correctly proposed by James Watson & Francis Crick.
- They constructed models to quickly visualise & assess viability of potential structures
- 1st model was triple helix with bases on outside & sugar-phosphate residues in centre, with Mg cross-links between strands.
- Guided by:
- Molecular distances & bond angles
– Linus Pauling - DNA composed of nucleotides made up of sugar + phosphate + base.
– Phoebus Levene -
DNA X-ray crystallography data showed it’s organised into helical structure – RF
(without permission):- DNA purified, then fibres were stretched in thin glass tube (to make most strands parallel)
- DNA targeted by X-ray beam, which diffracts when it contacted an atom
- Scattering pattern of X-ray distinctive
so recorded on film & used to find
DNA molecular structure: -
Structure:
- X in centre of diffraction pattern indicated DNA = double helix.
- DNA molecule shown to twist at regular intervals to form helix.
- X-shape angle showed pitch of helix.
- Orientation: N-bases closely packed together on inside & phosphates form an outer backbone
- Molecular distances & bond angles
- Rosalind Franklin rejected model as not enough Mg & didn’t support Erwin Chargaff’s findings (# of A + G = # of T + C).
- Using trial & error, Watson & Crick assembled DNA model that showed:
- DNA strands antiparallel & form double helix.
- DNA strands pair via comp. base pairing (A = T ; C Ξ G)
- Outer edges of bases remain exposed (allows access to proteins involved in transcription & replication)
- Potential DNA replication mechansims:
- Replication occurs via comp. base pairing (A pairs with T, G pairs with C)
- Replication is bi-directional due to antiparallel nature of strands
DNA Replication
Context:
- Occurs during S phase of Interphase.
- Replication is a semi-conservative process as new DNA contains 1 old & 1 new strand.
- Parent DNA strands act as templates for new
strands. - Comp. bases from opp. strands form H-bonds (A = T & C ≡ G)
- Occurs in a 5’ to 3’ direction.
- Consequently, when DNA replicated:
- Each new strand formed identical to original strand separated from template
- Both DNA molecules formed have equal
base seq. to original molecule.
DNA replication Process
- Helicase: Unwinds double helix by breaking H-bonds between 2 strands. → forms replic. fork with strands running in antiparallel directions.
- DNA Gyrase: ↓ strain created by helicase by relaxing + supercoils (via (–) supercoiling).
-
Single Stranded Binding (SSB) Proteins:
- Split DNA strands & prevent strand from re-annealing
- Prevent nucleases from digesting SS-DNA
- Dislodged from strand when new comp. strand synth. by DNA pol III.
- RNA Primase: Synthesises RNA primers on each template strand, RNA primers are binding spots for DNA-PIII.
-
DNA-PIII: Once bound to primer:
- Cleaves PO4’s from nearby DNsTP, to form nucleotides which are added to 3’ end of primer; according to comp. base pairing
(A = T + C ≡ G) - Cleaving PO4’s releases nrg used to form covalent bond between nucleotides.
- Thus, DNA-PIII synthesises new DNA between RNA primers (Okazaki frags).
- Cleaves PO4’s from nearby DNsTP, to form nucleotides which are added to 3’ end of primer; according to comp. base pairing
- Leading Strand: Con. synth. where DNA-PIII moves towards replic. fork.
- Lagging Strand: Discon. synth. where DNA-PIII moves away from fork → create Okazaki frags as fork exposes more temp. strand.
- DNA-PI: Excises RNA primers & replaces them with DNA;
- DNA ligase: Joins gaps between Okazaki frags by making cov. bonds between nucleotides to form a continuous strand.
Transcription
Sections of Gene:
- Promoter: Non-coding seq. that acts as binding site for RNA pol, thus starting transcription.
- Sense Strand: DNA strand with same base sequence as RNA. Not transcribed.
- Antisense Strand: Temp. strand with comp. base seq. to RNA & sense strand. Transcribed.
- Terminator: Non-coding seq. that signals RNA pol to detach from DNA, thus ends transcript.
Process:
- Transcription: RNA seq. synthesis using DNA template, by RNA polymerase. Occurs in nuc.
-
Initiation:
- RNA pol binds to promoter & unwinds + separates DNA strands by breaking H-bonds between comp. base pairs (using NRG).
- Nucleoside triphosphates (NsTP) line up opp. their comp. base partner.
-
Elongation:
- RNA pol excises 2 phosphates → NRG
- NRG used to bind (now) free nucleotides together (thus, synth. RNA) in 5’ → 3’ dir.
- (Forms coding seq.)
-
Termination:
-
RNA pol reaches terminator → both RNA pol & synth. RNA strand detach &
DNA double helix re-forms.
-
RNA pol reaches terminator → both RNA pol & synth. RNA strand detach &
Nucleosomes
- In eukaryotic organisms, DNA packaged with histones to form (nucleosome).
- Histones: Proteins used by cell to package DNA into nucleosomes.
-
Nucleosomes: Molecules consisting of 8 histones (octamer) with DNA coiled around.
- Help to supercoil DNA, resulting in greatly compacted structure that allows for more efficient storage.
- Histone tails are + charged, so associate to DNA & determine tightness of packing
-
Acetylation: Adding acetyl group to histone tail, dec. + charge → DNA less tightly coiled & inc. transcript.
(Euchromatin) -
Methylation: Adding methyl group to histone tail, inc. + charge → DNA more tightly coiled & dec. transcript.
(Heterochromatin)
-
Acetylation: Adding acetyl group to histone tail, dec. + charge → DNA less tightly coiled & inc. transcript.
- Supercoiling helps:
- Protect DNA from damage
- Mobilise chrom’s during mitosis & meiosis.
Organisation of Eukaryotic DNA
- DNA packaged with 8 histone proteins (an octamer) → complex (nucleosome)
- H1 histone binds to linker DNA, which binds nucleosomes together, to form
* *chromatosomes** - These coil to form more condensed solenoid fibre structure, which then form loops.
- Loops compressed & folded around protein scaffold to form chromatin (eu- or hetero-).
- Chromatin then supercoils during cell division to form chromosomes visible (when stained) under microscope
Epigenetics
Epigenetics: Study of phenotypic changes caused by variations in gene expression levels.
- DNA Methylation prevents TF binding, so ↓ gene expression/transcription.
- Thus, transcription of gene ind. prop. to DNA methyl.
- Epigenetic analysis shows that DNA methylation patterns/gene expression may change over course of a lifetime
- Diff cell types in same organism may have markedly diff DNA methylation patterns.
- Env. factors (e.g. diet, pathogen exposure, etc.) also affect DNA methyl. within cells.
- Also influenced by heritability but not genetically pre-determined, so identical twins may have diff DNA methyl. patterns.
Transcription Regulation
- Transcription regulated by 2 groups of proteins that mediate binding of RNA pol to promoter:
- Transcription factors (TF) form complex with RNA pol at promoter & don’t allow initiation without factors, hence their levels regulate gene expression.
-
Regulatory proteins bind to non-coding DNA seq. outside of promoter & interact with transcription factors:
- Activator proteins bind to enhancer
seq. & ↑ transcript. rate
(by mediating complex formation) - Repressor proteins bind to silencer
seq. & ↓ transcription rate
(by preventing complex formation)
- Activator proteins bind to enhancer
-
Control Elements: Exist in large amounts to further tighten control & coordination.
- Distal control elements bind to regulatory proteins.
- Proximal control elements bind to transcription factors.
- Presence of certain transcription factors or regulatory proteins may be tissue-specific
- Intracellular chem signals may also trigger change in [reg. prots] or [TF] in response to stimuli → gene expression changes in response to changes in conditions in/out cell:
- Humans produce different amounts of melanin depending on light exposure
- Morphogens: Uneven distr. in embryo & contribute to diff. gene express. patterns depending on their conc.
- Mutant allele “cs” in “C” gene in Siamese cats only produces tyosinase (pigment production) at < body temp.
- In eukaryotes, post-transcriptional mod. of transcript mRNA needed to form mature mRNA
- Ribosomes also separated from genetic material (DNA & RNA) by nucleus, so gen. needs to be moved.
3 post-transcriptional events:
-
Capping: Involves addition of methyl group to 5’-end of transcribed RNA
- Methylated cap provides protection against degradation by exonucleases
- Allows transcript to be recognised by ribosome.
-
Polyadenylation: Addition of poly-A tail to 3’-end of mRNA. (NOTE: A stands for adenine)
- Poly-A tail improves RNA transcript stability
- Facilitates its export from nucleus.
- Thus, mRNA mods allow ribosome to it out of nuc. (via nuclear pores) before transl.
-
Splicing: Removing introns from mRNA transcript.
- Within eukaryotic genes exist:
- Introns: Non-coding seq. which must be removed prior to forming mature mRNA.
-
Exons: Coding regions, which fuse
together when introns removed to form continuous seq.
-
Alternative splicing: Removing specific
exons → Gene seq. make diff polypep’s.
- Within eukaryotic genes exist:
-
Polysome: Group of ribosomes translating an mRNA seq. simul.
- In prok. they couple transl. + transc. due to no comp. & both occuring in 5’ → 3’.
Translation
Context:
- Translation: Polypeptide synthesis using base sequence of mRNA molecules (in ribosomes).
- mRNA seq. read by ribosome in base triplets (codons). Each codon codes for 1 AA.
- Gen code degenerate as >1 codon can code for same AA. Also allows silent mutations to occur, whereby a change in DNA seq. doesn’t alter polypeptide seq.
- Thus, order of codons in mRNA seq. determine AA order in polypeptide chain.
- 64 codon possibilities (4 bases 3 bases/codon)
- 3 components work together to synthesise polypeptides by translation:
- mRNA has sequence of codons that determines AA sequence of polypeptide.
- tRNAs have anticodons that bind to comp. codon on mRNA; they carry AA corresponding to that codon.
- Ribosomes are mRNA & tRNA binding sites; also catalyse polypeptide assembly.
Process:
- Initiation: Assembly of components that carry out translation (mRNA, tRNA, ribosome).
- Next, appropriate tRNA molecule binds to
codon via its anticodon (according to comp. base pairing) - Finally, large ribosomal subunit aligns itself to the tRNA molecule at the P site and forms a complex with the small subunit
- Ribosome composed of 2 sub-units:
- Small subunit binds to mRNA & moves along it until reaching start codon (AUG).
-
Large Subunit containing 3 tRNA binding sites: A, P, and E binds to small subunit.
* Elongation:
- Initiator tRNA (with methionine) binds to start codon “AUG” in P site of large sub.
- 2nd tRNA (with anticodon comp. to 2nd codon) binds to 2nd codon in A site of large subunit.
(max 2 tRNAs bound at once). - Ribosome catalyses pep bond between AAs in A & P site via condensation reactions → dipep.
- tRNA in P site now deacylated (no AA), whilst
tRNA in A site carries dipeptide. - Ribosome translocates 3 bases along mRNA in 5’ to 3’ dir, so 1st tRNA moves from P to E site, releasing it.
- 2nd tRNA takes place of 1st. so moves from A to P site, freeing A site.
- 3rd tRNA binds with anticodon comp. to 3rd
codon on mRNA in vacant A site. - Process repeated until stop codon is reached.
- Termination: Disassembly of components & release of polypep chain.
- Non-coding stop codon reached, which release factor signalling transl. end; polypep released.
- Ribosome disassembles back into its 2 indep. subunits.
Activating tRNA
Context:
- tRNA activation occurs in cytoplasm via tRNA-activating enzyme, tRNA, AA and ATP.
- Each AA recognised by specific enzyme
- But multiple tRNA molecules recognised by enzyme due to degeneracy.
Process:
- tAE binds to specific AA & ATP.
- Enzyme catalyses ATP hydrolysis → AMP + 2P
- AA binds to AMP → AA-AMP complex, linked by high energy bond, 2P is released.
- Bond act as energy store to provide most of
nrg used to make pep. bond during transl. - tRNA binds to tAE.
- AA cov. bonded to 3’ terminal of tRNA, releasing AMP attached to enzyme.
- tRNA molecule now “activated” & released.
Insulin production in bacteria
Context:
- Diabetes type II due to destruction of ß-cells that secrete insulin (hormone).
- Used to be treated with insulin produced from other animals (e.g. pigs) as they bind to human insulin receptor, but found to cause allergies.
- Genetic code = universal as same codons code for same AAs in all living things, gen. info
transferrable between species - Ability to transfer genes between species used to produce human insulin in bacteria (for mass production), with exactly same AA seq. as gene
transcribed & translated in human cells.
Process
- Desired gene seq. obtained by either:
- DNA isolated from cells & nuclei by centrifugation (heavy cell organelles sink).
- Using rev. transcriptase to convert mRNA → dDNA.
- Interest gene specifically amplified via PCR
- Plasmids used as vectors as they can auton. self-replicate & express genes
- Gene + plasmid cut with same restriction endo.
at specific recognition sites by cleaving sugar-phosphate backbone to create “sticky ends” - Gene now binds to plasmid as sticky ends of gene & vector overlap via comp. base pairing.
- Gene & vector spliced together by DNA ligase (which fuses their backbones together with phosphodiester bond) to form recomb DNA.
- Recomb. DNA introduced into host cell/org.
(transfection if prok or transformation if euk). - Antibiotic selection commonly to ID which cells have successfully incorporated recomb. DNA.
- Transgenic cells, once isolated & purified, express desired trait encoded by int. gene, so placed in fermenter to reproduce lots.
- (e.g. Insulin) produced, purified and sold for use (e.g. in diabetics).
Hershey-Chase Experiment
- Proteins & nucleic acids believed to be involved in composition of genetic material.
- Alfred Hershey & Martha Chase conducted a series of experiments to prove DNA was gen material (not protein).
- Known that viruses (E.g. T2 Bacteriophage)
consisted solely of DNA & protein coat and could transfer their genetic material into hosts. - T2’s were grown in 1 of 2 isotopic mediums in order to radioactively label specific viral part.
- Viruses grown in radioactive S (35S) had radiolabelled proteins (S present in proteins but not DNA)
- Viruses grown in radioactive P (32P) had radiolabelled DNA (P present in DNA but not proteins)
- Viruses allowed to infect bacterium (E. coli).
- Virus & bacteria separated via centrifugation
- Larger bacteria forms solid pellet whilst smaller viruses remains in supernatant
- So when pellet found to be radioactive when infected by 32P–viruses (DNA) but not 35S–viruses (protein), showed DNA passed on.
- Showed DNA, not protein, was gen material as DNA was transferred to bacteria.
Meselson and Stahl
- Prior to experiment, 3 hypotheses had been proposed for the method of replication of DNA:
- Conservative Model: Entirely new molecule is synthesised from a DNA template (which remains unaltered)
- Semi-Conservative Model: Each new molecule consists of 1 newly synthesised strand & 1 template strand
- Dispersive Model: New molecules made of segments of new & old DNA
- Meselson & Stahl experimentally tested validity of these models using N15 (heavier radioactive isotope of N14), an element present in bases).
- DNA cultured in N15 for many gens to ensure N15 was only N source in DNA, then transferred to, & induced to replicate in N14-only medium.
- DNA samples separated via centrifugation to find DNA composition in replica. molecules.
- DNA detected as it absorbs UV, hence creating dark band when tubes illuminated with UV:
- Single band in 1st gen falsifies cons. replica. (shows mix of old & new DNA/
N15 & N14). - 2 bands in 2nd gen falsifies dispersive replication (New-only & mixed DNA /
Only-N14 and N15 & N14).
- Single band in 1st gen falsifies cons. replica. (shows mix of old & new DNA/
- Hence, showing DNA Replication is semi-conservative.
DNA Sequencing
- DNA sequencing: Process by which base order of a nucleotide sequence is elucidated
-
Dideoxynucleotides (ddNT): Lack 3’-OH
group needed for making PPD bond.- Thus, ddNTs stop further elongation of nucleotide chain & effectively end replica.
- Resulting DNA seq. reflects specific nucleotide pos. ddNT was added.
-
Sanger Method:
- PCR mixes set up, each containing stocks of deoxyribonucleotides + ddNT + fluoresc. primers + enzymes for replica.
- PCR makes lots of DNA molecules quickly, so PCR mixes should have all possible terminating frags for that spec. base.
- Frags separated using gel electroph & base seq. determined by ordering frags according to length.
- Fuorescently labelled primer included in each mix allow frags to be detected by automated seq. machines.
- If Sanger method conducted on coding strand, resulting seq. elucidated will be identical to template strand.
Non-coding DNA
- Vast majority of human genome is comprised of non-coding DNA, which serve other functions (table).
Context:
- DNA profiling: Technique by which individuals identified & compared via resp. DNA profiles
- Within non-coding regions of individual’s genome exists satellite DNA: DNA seq. made up of repeating elements (STRs).
Process:
- DNA sample collected (e.g. from blood, semen, saliva, etc.), then amplified using PCR
- Sat. DNA (with STR seq.) cut with specific restriction endo. to create frags, which differ between indivs due to # of STRs in frags.
3. Frags separated using gel electrophoresis & resulting profiles (composed of bands) are compared to see if bands match.
-
For family tests:
- Paternal lineage determined by analysing VNTR from Y-chromosome.
- Maternal lineage deduced by analysing mitochondrial DNA variations in single nucleotides at hyper-variable regions.

Genes, Locii, Alleles, Chromosomes, Homologous Chromosomes, SNPs, Mutations,
- Polygenic traits: Traits influenced by multiple genes.
- Gene: Heritable factor consisting of a length of DNA and influences a specific characteristic.
- Locus: Specific position on chrom occupied by gene.
-
Allele: Alt. forms of same gene, they have same locus, but only one can occupy it.
- Code for diff. variations of specific trait
- Alleles have very similar gene seq. & only diff. by 1 or few bases (SNP’s).
- Single Nucleotide Polymorphisms (SNP): Positions in gene where >1 base may be present.
-
Chromosomes: Groups of linked genes.
23 types of chromosomes in humans.- Homologous: Chromosomes that carry same sequence of genes but not necessarily same alleles of those genes, allowing species members to interbreed.
-
Mutations: Random changes in base seq. of gene. May lead to new alleles forming from other alleles.
- Base Substitution: 1 base in seq. of gene replaced by different base.
- Almost all mutations either neutral or harmful as random change to allele selected for by NS over time unlikely to be beneficial.
- Mutations in body cells eliminated when individual dies, but mutations in sex cells can be passed on to offspring & cause genetic disease.
Mutations
- Gene mutation: Change in nucleotide/base seq. of gene coding for a specific trait.
- New alleles are formed by mutation
(alleles only differ by few bases (SNP’s)). - Either spontaneous or induced.
Gene mutations can be:
- Beneficial: Change gene seq. (missense mutations) to create new variations of a trait
-
Detrimental: Truncate gene seq. (nonsense
mutations) to STOP normal function of trait. - Neutral: Have no effect on functioning of specific trait (silent mutations).
Induced by:
- Ionising Radiation ↑ mutation rate if enough nrg to cause chemical changes in DNA.
-
Chemical Mutagens ↑ mutation rate by causing chemical changes in DNA.
(e. g. mustard gas, benzene, ROS, tar) - Biological: Viruses (HPV) and Bacteria
Hiroshima + Nagasaki and Chernobyl

Sickle Cell Anaemia
- Disorder caused by single base sub. mutation to a gene (Hb), which codes for haemog. prod.
- Most humans have co-dom. allele Hb4.
-
DNA: CTC → CAC on 6th codon of gene
→ new co-dom. allele formed: Hb5. - mRNA: GAG → GUG.
- Polypeptide: 6th AA in haemog: Glu → Val when GUG transcribed.
- Mutation → changes haemog. structure →
stick to form insoluble fibrous strands → rigid enough to distort RBCs into sickle shape. -
Consequences:
- Damage to tissues by becoming trapped in & blocking blood capills → ↓ bloodflow.
- Both haemog & plasma memb. dmg
- RBC Life ↓ → ↓ RBC count → Body can’t replace RBC at same rate → anaemia.
Genome + HGP (PM MS MEGI)
Genome: Whole of gen. info of organelle, cell, or organism. Includes genes + non-coding DNA seq.
- In animals, genome = DNA molecules that form chromosomes in nucleus + DNA molecule in mitochondrion.
- 46 chromosomes in humans.
- In plants, genome = DNA molecules that form chromosomes in nucleus + DNA molecules in mitochondrion + chloroplast.
- In prokaryotes, genome = DNA in circular chromosome + any plasmids that are present. Much smaller as a result.
- Genome size generally ∝ size:
- Viruses & bacteria tend to have smallest genomes
- Plant genome size varies dramatically due to capacity for plant species to self-fertilise & become polyploid.
- Size may also change due to chromosomes fusing or splitting, but rare.
HGP: International cooperative venture established to sequence human genome
- HGP showed that humans share majority of their seq, with SNP’s contributing diversity.
- HGP aided by improvements in tech that rapidly inc. speed of gene sequencing.
- Completion of HGP led to many outcomes:
- Mapping: #, location, size & seq. of human genes now established
- Screening: Allowed for prod. of specific gene probes to detect sufferers & carriers of genetic diseases.
- Medicine: Discovery of new proteins & causes of gen. diseases → ↑ treatments.
- Evolution: Comparing with other genomes → ↑ knowledge of origins, evolution & migratory patterns of man
- Gene transfer (genetic engineering)
- Promote International co-operation.
- Understanding that genome > proteome & that most genome not transcribed.
- Mutations discovered.
(e.g. introns, promoters, STR’s, etc.)
Euk + Prok DNA + Plasmids
-
Plasmids:
- Contain few genes
- Capable of self-replication
- Exchanged between bacteria via pili (conjugation) → bacteria evolves new features within gen (horiz. gene transfer).
- Plasmids may also cross species barriers if plasmid released when prok. absorbed by cell of different species.
- Plasmid’s ability to self-replicate & synth. proteins → vectors for genetic engineering.

Homologous Pairs
- Sexually reproducing organisms inherit their genetic seq. from both parents
- So organisms possess 2 copies of each chrom (homologous chrom), which share:
- Same structural features (e.g. same size, same banding patterns, same centromere positions)
- Same genes at same loci positions (whilst
genes are same, alleles may be different) - Homo. chrom. separated in gametes (via meiosis) prior to reproduction, in order to prevent chrom. numbers continually doubling with each generation.
- Organisms with diff. diploid #’s unlikely to be able to interbreed (can’t form homo. pairs in zygotes).
- In cases where diff. species do interbreed, offspring usually infertile (can’t form functional gametes). (e.g. horse + donkey).
Karyograms
-
Karyotypes: # & types of chrom. in euk:
- Harvest cells (usually from foetus or adult WBC’s).
- Cell div chemically induced, then mitosis arrested whilst chrom. are condensed
(so they’re visible). - Stage during which mitosis halted determines whether chrom. appear with sister chromatids or not.
- Chroms stained & photo taken, then
arranged into homo. pairs by size
(sex chrom. shown last) → karyogram.
-
Karyotyping usually done prenatally to:
- Determine gender of unborn child (via ID of sex chrom.)
- Test for chromosomal abnormalities (e.g. Down)
-
Down syndrome:
- Due to non-disjunction in 1 of parental gametes: Failure of chrom separation resulting in 1 xtra/1 less chrom.
- Non-disjunction may occur via:
- Sis chromatids in Anaphase II
→ 2 affected daughter cells - Bivs failing to separate in Anaphase I → 4 affected daughter cells.
- Sis chromatids in Anaphase II
- Down = Trisomy due to: 1 parental gamete having 2 chrom 21 copies (due to non-disj) + 1 normal parental gamete with 1 copy fusing → 3-copy zygote.
- Studies show that:
- Non-disj ∝ parental (esp. mom) age ↑
- May be due to developing oocytes being arrested in prophase I until ovulation as part of oogenesis.
- Higher incidence of chrom. errors in offspring due to anaphase I non-disjunction.
- Mean maternal age ↑, → ↑
# of Down syndrome offspring.
- Non-disj ∝ parental (esp. mom) age ↑
-
Karyotyping sources:
- Amniocentesis: Extraction of amniotic fluid (has fetal cells) with needle inserted through abdomen.
-
Chorionic Villus Sampling (CVS):
Extraction of CV (placental tissue) with suction tube inserted through cervix. - CVS can be done earlier in preg. than
Amniocentesis, but risk = 2% (opp. to 1%).
Autoradiography
John Cairn 1st to create image of chroms before condensation, which made measuring inaccurate.
- Incubate cells in radioactive 3H-T solution.
- 3H-T incorporated into chromosomal DNA of cell as T not present in RNA.
- Chromosomes isolated by gently lysing cells & fixing chrom. to photographic surface
- Surface then immersed in radioactively-sensitive emulsion containing AgBr.
- Radiation released from 3H-T converts Ag+ ions in AgBr into insoluble Ag grains.
- Following period of exposure, excess AgBr
washed away, leaving Ag grains = small black • - When photographic film developed, chrom. DNA visualised with an electron microscope.
Meiosis
Meiosis: Reduction div of diploid germline cells in reproductive organs into 4 gen. distinct hap nuclei (gametes)
- 1st meiotic division separates homo. pairs to halve chrom. # (diploid → haploid) (red. div).
- 2nd meiotic division separates sister chromatids
-
Interphase:
- DNA replic. during S phase so red. div. → ½ diploid # of chroms in gametes.
- Thus, when gametes fuse during fertilisation, they form diploid zygote.
- If not, polyploidy occurs.
- As meiosis results in gen distinct gametes, random fert. by egg & sperm always generates diff. zygotes
-
Prophase I:
- Synapsis: Homo. chromosomes pair up at points (chiasmata) to form bivs (synapsis)
- Crossing-over: DNA exchanged between non-sister chromatids across chiasmata → chromatid alleles recomb.
- Chiasmata: Inc. stability of bivalent, so likely occcur at random pos &/or >1 in each, so cross-over can occur anywhere.
-
Metaphase I:
- Random Orientation: Orientation of bivs is random.
-
Anaphase I:
- Disjunction: Splitting of homologous chroms to opp. poles.
-
Random & ind. orientation → random & ind. assortment → Gamete combos = 2n
(n = haploid number). - If crossing over also occurs, gamete combos becomes immeasurable

Gregor Mendel’s Laws
- Gregor Mendel came up with inheritance laws by performing experiments on pea plants
- He crossed diff. purebred pea plant varieties.
- Collected & grew their seeds to determine their traits.
- Next, he self-fertilised offspring & grew their seeds to similarly determine their traits.
- Crosses were performed many times to establish reliable data trends.
- Mendel discovered the following things:
- When crossing 2 diff. purebred varieties together, results weren’t a blend – only 1 feature expressed
- When Mendel self-fertilised offspring, 2 diff traits expressed in 3:1 ratio.
- From these findings, Mendel drew the following conclusions:
- Organisms have discrete factors (genes) that determine its features.
- Furthermore, organisms possess 2 versions (alleles) of each factor (gene).
- Each gamete contains only 1 allele (hap).
- Parents contribute equally to inheritance of offspring due to fert. of randomly selected egg & sperm
- Certain laws were derived:
- Law of Segregation: When gametes form, alleles separated so that each gamete carries only 1 allele for each gene.
-
Law of Independent Assortment: The segregation of alleles for 1 gene occurs independently to that of any other gene*
- * Not true for linked genes.
-
Principle of Dominance: Recessive alleles will be masked by dominant alleles.*
- * Some genes show co-dom.
Genotype, Phenotype, Dominant, Recessive, Co-dominant
- Genotype: Allele combo for specific trait.
-
Phenotype: Observable traits of a specific trait.
(determined by genotype & env. influences). - Dominant: Expresses trait that is always present in phenotype when present.
- Recessive: Only expressed in phenotype when in a homozygous state.
- Co-dom: Allele pairs that affect phenotype equally when present in heterozygote.
ABO Blood groups
- Human RBCs categorised into blood groups based on structure of its antigen.
- IA, IB & i alleles all produce basic antigen on surface of RBCs:
- IA & IB alleles co-dom & each modify antigen to produce different variants.
- i allele is recessive & doesn’t modify the basic antigenic.
- Incorrect blood transfusions lead to agglutination and lysis of RBCs.

Genetic Disorders
- Sex Linkage: Gene controlling trait located on sex chrom. Most X-linked as ↓ genes exist in Y.
-
Sex Rules:
- Only ♀ carriers as ♂ can’t be hetero.
(only have 1X). - ♂ always inherit X-linked trait from mom
(as Y received from dad). - ♀ can’t inherit X-linked recessive trait from unaffected dad (must receive dom allele).
- Only ♀ carriers as ♂ can’t be hetero.
- Many genetic diseases identified, but most are rare because:
- Alleles that ↓ survival + reprod. unlikely to be passed onto offspring.
- Recessive conditions more common, as faulty allele can be present in carriers without causing disease/harm.
- Dominant conditions may have late onset, as this doesn’t prevent reprod. & the transfer of faulty allele.

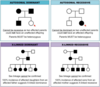
Pedigree Charts
PCR
-
PCR: Artificial DNA replic. technique used to
amplify specific DNA seq. - Useful when only small amount of DNA available for testing. E.g. crime scene samples of blood, semen, tissue, hair, or from fossils
- Reaction occurs in thermal cycler & uses variations in temp to control replic:
-
Denaturation: DNA sample heated (90ºC) to break H-bonds holding strands tog. →
separates them into single DNA strands. - Annealing: Sample cooled (50ºC) to allow primers to anneal to target DNA seq. + re-form double strand apart from seq. with primers (target seq).
-
Elongation: Sample heated to opt. temp.
for heat-tolerant Taq polymerase (doesn’t denature at high temp) to function (75ºC), isolated from thermophilic bacteria.
-
Denaturation: DNA sample heated (90ºC) to break H-bonds holding strands tog. →
- Repeat procedure. For n cycles, PCR produces 2n copies of DNA sample.
Gel Electrophoresis
- Gel Electrophoresis: Used to separate charged molecules, like proteins or DNA fragments, according to their size and charge:
- Fragmented DNA samples placed in gel block & current applied → samples move through gel
- Small fragments less impeded by gel so move faster than large fragments, so orders by size.
DNA Separation:
- DNA fragmented with restriction endonuclease - diff. DNA samples generate diff frag lengths.
- DNA samples placed into agarose gel &(DNA)¯ due to (PO43¯) so moves to anode, but at diff. rates.
- Frag sizes calculated by comparing against known industry standards.
- Separated seq. moved to membrane &
specific seq. ID’d by adding comp hybridisation probe, which appears in autoradiograph.
Protein Separation:
- Proteins may fold into diff shapes/ diff. sizes & have + & – regions, so treated with anionic detergent (SDS) to induce uniform – charge.
- Protein samples placed into polyacrylamide gel & move to anode at diff. rates dep. on size.
- Protein sizes calculated by comparing against known industry standards.
- Separated proteins moved to membrane &
target proteins identified by staining with specific monoclonal antibodies.
GMO’s

Bt corn
- Bt corn: GM maize with insecticide-prod. gene (Bt)
- Bt-toxin kills corn borers, which eat crop
- Monarch butterflies also feed on milkweed with Bt-corn pollen moved by wind.
Experiments:
- 1st Experiment conducted comparing monarch caterpillars DR’s & Bt pollen-based diets.
- Monarchs fed milkweed leaves with Bt
pollen (sim. spread via wind). - Growth & DR’s compared in monarchs fed on non-dusted, non-GM & dusted diets.
- Caterpillars exposed to Bt pollen ate less, grew more slowly & exhibited higher DR.
- Monarchs fed milkweed leaves with Bt
- But, Bt pollen on leaves > found naturally (rain could wash) & Larva restricted in diet (in field, larva could avoid eating pollen dusted leaves).
- 2nd experiment conducted comparing monarch butterfly DR’s & proximity to Bt corn fields:
- No sig. ↑ DR when monarch larva placed in or near actual Bt corn field
- Exposure to Bt pollen poses no sig. risk to monarch butterfly pops.
Clones
- Clones: Groups of gen. identical organisms, derived from single original parent cell.
Natural Methods of Cloning:
- Bacteria & fungi reproduce asex. to produce genetic clones via binary fission (mainly bacteria) or by producing spores (mainly fungi)
-
Vegetative propagation: Small pieces induced to grow indep. due to totipotent meristematic tissue in adult plants differentiating:
- Onion/garlic bulbs = modified plant leaves – all bulbs in group are gen. identical
- Underground stems (e.g. potato tubers) can form new plants gen. identical to parent plant
- Some animal species also reproduce asexually:
-
Binary Fission: Parent organism divides equally → 2 daughter organisms
(e.g. flatworms) -
Budding: Cells split off parent organism, creating smaller daughter organism that
eventually separates from parent (Hydra) -
Fragmentation: New organisms grow from separated frag of parent org
(e.g. starfish) - Parthenogenesis: ♀ prod. diploid egg cells instead of haploid (e.g. ♀ aphids)
-
Binary Fission: Parent organism divides equally → 2 daughter organisms
-
Human Twins:
- Monozygotic (ID) twins created when fertilised egg splits into 2 identical cells, each forming an embryo. Gen. identical.
- Dizygotic (non-ID) twins created when unfertilised egg splits into 2 cells & each fertilised by diff. sperm.
Artificial Methods of Cloning
Embryonic Division:
- Embryonic cells retain pluripotency, so
differentiate to form all tissues comprising org. - Embryonic cells separated artificially in lab, early in developmental cycle.
- Separated groups of cells implanted into surrogate uterus to develop into clones.
- Limited by fact that embryo used still formed randomly via sexual reproduction & so specific genetic features of resulting clones unknown.
(SCNT): Artificial method by which cloned embryos prod. using differentiated adult cells.
- Genetic features of resulting clone known.
- Somatic cells taken from adult donor & their cultured (for their diploid nuclei).
- Enucleated egg cell made by taking unfertilised egg & removing its nucleus.
- Egg fused with somatic nucleus → diploid egg.
- Electric current used to stimulate egg to divide & develop into embryo.
- SCNT cloning split into 2 purposes:
- Reproductive cloning: If embryo is implanted into surrogate uterus, new cloned organism of donor will develop.
-
Therapeutic cloning: Embryonic cells
induced to differentiate to create specific tissues or organs for transplantation

Stem Cuttings (MELT HAP)
- Stem cutting: Separated portion of plant stem that can regrow into new indep. clone via vegetative propagation.
- All stems possess nodes, from which leaf, branch or aerial root may grow.
- Stem cuttings typically placed in soil with lower nodes covered & upper nodes exposed
- Stem cutting: Common method employed to rapidly propagate plant species (including sugar cane, grapes & roses)
- Factors affecting rooting of stem cutting:
-
Cutting pos: Cutting stem above/below
node & relative proximity of cut to node) - Cutting length: How many nodes remain on cutting.
- Growth medium: Soil, H2O, potting mix, compost, or open air
- Use & conc. of growth hormones
- Temp: Most cuttings grow optimally at temps common to spring & summer
- H2O availability: Groundwater or humidity
- Soil pH:
- Light Exposure:
-
Cutting pos: Cutting stem above/below
Thomas Morgan
- Thomas Morgan discovered non-Mendelian ratios in fruit flies, which aided in understanding gene linkage.
- Cross-breeding R-eyed wild types with W-eyed mutants → clear sex bias in phenotypic distr.
- All ♀ offspring of R-eyed ♂ were R-eyed,
- All ♂ offspring of W-eyed ♀ were W-eyed.
- Morgan inferred that eye colour gene dep. on X, as it was found on X-chromosome.
- Morgan then investigated other traits & found that certain phenotypic combos occurred in much lower freq. than expected.
- Based on this data, Morgan proposed:
- Alleles for Link-traits located on same
chrom (link-genes), so didn’t indep. assort. - Linked alleles could be uncoupled via cross-over to create alt pheno, but new
phenotypes would occur at lower freq. - Morgan also observed that cross-over freq. between linked genes differed depending on distance between 2 genes on chrom (cross-over freq. ∝ distance between genes). Which he used to dev 1st gene linkage maps, showing relative positions of genes on chrom.
- Alleles for Link-traits located on same
Chi-Squared Test
- Offspring with unlinked genes inherit any potential phenotypic combo equally
(due to random assortment).
of alleles (due to independent assortment). - Offspring with linked genes only express the phenotypic combos present in either parent (unless crossing over occurs)
- Thus, unlinked recomb. phenotypes occur less freq. than ‘linked’ parental phenotypes.
Chi-squared: Statistical measure used to determine whether diff. between obs. & exp. freq. distribution is statistically significant.
-
Idenify Hypotheses:
-
H0: No sig. diff. between obs. & exp. freqs
(i. e. genes are unlinked) -
H1: Sig. diff. between obs. & exp. freqs
(i. e. genes are linked)
-
H0: No sig. diff. between obs. & exp. freqs
-
Construct freq. table:
- Draw a dihybrid cross to find exp. ratios.
- Total x exp. ratio = exp. freq
- Apply Chi-Squared: (O - E)2 ÷ E
- Degrees of Freedom: (m - 1)(n - 1) = 3 for dihybrid crosses.
-
ID p value:
- When df = 3, Chi-squared > 7.8 to be considered stat. sig. (p < 0.05).
- If > 7.8 → p < 0.05 → Sig. diff
→ genes are linked. - If < 7.8 → p > 0.05 → Sig. diff.
→ genes are unlinked.
Polygenic Inheritance
- Monogenic traits (controlled by 1 gene loci) exhibit discrete variation, with individuals expressing 1 of several distinct phenos.
-
Polygenic traits (controlled by >2 gene loci) exhibit continuous variation, with individuals
expressing pheno existing in a bell-shaped cont. spectrum of potential phenos.- # of loci responsible for particular trait ∝ # of possible phenos.
-
Maize grain colour: Controlled by 3 gene loci:
- Grain colour ranges from W to dark R, dep. on amount of pigment expressed
- Each gene has 2 alleles, which either code for R or W pigment.
- Most freq. combos have equal # of both.
- Conversely, combos of extremes are rare
- Overall pattern of inheritance shows continuous variation.
-
Height + Skin Colour also affected by env:
- Added effect of env pressures functions to ↑ variation seen for particular trait
- Human height controlled by mutliple genes, but also affected by diet & health (disease).
- Skin colour controlled by multiple melanin producing genes, but also affected by sun exposure.
Evolution by Natural Selection (ICE AGE)
- Evolution: Cumulative change in allele freq. of a pop’s gene pool over successive gens.
- Gene pool: Sum total of alleles for all genes present in a sexually reproducing population
- Gene pool size ∝ amounts of gen diversity → ↑ chances of biological fitness & survival
-
Natural Selection: Freq. of alleles that adapt
indivs to env. ↑, + vice-versa to bad alleles.
Process (ICE CAGE)
- Inherited variation exists within population
- Competition ← offspring > env. capacity.
- Env pressures → differential reprod. within pop
- Adaptations (traits that make indiv. suited to its env & way of life) that benefit survival select for and passed onto offspring.
- Genotype/Allele freq. changes cumulatively within pop gene pool across gens (Evolution).
Types of selection
-
Stabilising Selection: Natural sel. favours intermediate pheno over both ends of the range of variation.
- Results in removal of extreme phenotypes (phenotypic distribution becomes centrally clustered to reflect homogeneity)
- Operates when env conditions are stable & competition is low.
- E.g. Human birth weights (too large = birthing complications ; too small = risk of infant mortality)
-
Directional Selection: Natural sel. favours 1 end of the range of variation over another.
- Operates in response to gradual or sustained changes in env conditions
- Causes progressive change in pop in that direction.
- Causes species to change enough over time to be regarded as different species (speciation).
- E.g. Dev of antibiotic resist in bacteria pop
-
Disruptive Selection: Natural sel. favours both ends of the range of variation at cost of
intermediate phenos - Causes pheno distr to deviate from centre & results in bimodal spread
- Occurs when fluctuating env conditions (e.g. seasons) favour presence of 2 diff
phenos, which are adapted to diff. niches. - Extreme types adapted to diff. niches
(e. g. seasons). - Reproductive barriers become established between extreme types (e.g. plants grow in diff. seasons)
- E.g. proliferation of black or white moths in regions, but not grey-

Change in allele freq.
- Population bottlenecks & founder effect will exacerbate genetic diffs between geographically isolated pops.
-
Pop bottlenecks: Natural or anthropomorphic event that ↓ pop size by an order of magnitude (~ >50%).
- Surviving pop has less genetic variability than before, so subject to higher lvl of genetic drift.
- As surviving members begin to repop, newly deving gene pool div to original.
-
Founder effect: Occurs when small pop breaks away from larger pop to colonise new territory
- As this pop subset doesn’t have same degree of diversity as larger pop, it’s subject to more genetic drift
- Consequently, as this new colony ↑ size, its gene pool diverges from original.
- Original pop stays largely intact as opposed to pop bottlenecks.
Evidence for Evolution
Fossils:
- Fossils show changes over time in organisms; as fossilised orgs are diff. from existing ones; yet share homo structures with existing orgs;
- Seq. of fossils at various geological eras matches expectations of evo:
- Bacteria
- Simple invertebrates
- Complex vertebrates.
- They also show intermediate stages in evolution of groups.
- Thus suggest orgs share common ancestors.
- Some have not changed much, due to little selection pressure. There are also gaps in fossil record, which make conc. difficult.
Selective Breeding: Form of artificial selection whereby humans select for desirable traits to be passed onto future gens (rather than env).
- Modern varieties of wheat & rice prod. higher yields & more pest-resist. than wild ancestors.
- Dog breeds are numerous and sig. different from wild wolf ancestor.
- Artificial selection shows changes in domestic species can be achived in relatively short time.
- Does not prove that evolution of new species has occured.
Homo. structures: Same ancestor that had this structure, but has become diff. as they perform different functions. Implying common ancestry.
- The more similar the homo. structures between 2 species are, the more closely related they are likely to be.
- E.g. pentadactyl limbs in all vertebrae:
- Human hands adapted for tool manipulation.
- Wings adapted for flying
- Hooves adapted for galloping
- Fins adapted for swimming
- AA seq. of diff. species also show how closely related species are.
- Speciation:
- Provides evidence for evolution of species & origin of new species by evolution due to continuous range in variation between populations.
- But doesn’t match either belief that:
- Species created as distinct types of organism, so should be constant across their geographic range
- Species are unchanging.
Fossils
-
Fossil: Preserved remains or traces of any organism from the past.
-
Reserved Remains: Provide direct
evidence of ancestral forms.
(e.g. bones, teeth, shells, leaves, etc.) -
Traces: Provide indirect evidence of ancestral forms.
(e.g. footprints, tooth marks, burrows)
-
Reserved Remains: Provide direct
- Fossil Record: Totality of fossils, both discovered & undiscovered.
- Fossil record provides evidence for evo. by revealing features of ancestor for comparison against living descendants.
Law of Fossil Succession: Chron. seq. by which traits appear to develop.
- Fossils dated by determining age of rock layer in which fossil is found (rock layers develop in chronological order so oldest = bottom).
- Different kinds of organisms found in rocks of particular ages in consistent order, indicating a seq. of dev:
- Prokaryotes appear in fossil record before eukaryotes
- Ferns appear in fossil record before angiospermophytes.
- Invertebrates appear in fossil record before vertebrates
- Ordered succession of fossils suggests that newer species likely evolved as a result of changes to ancestral species
Industrial Melanism
- Inherited variation exists within pop (mel. + pep.)
- Comp. ← offspring > env. capacity.
- Env press (poll due to ind. revo.) → SO2 kills pale lichen + C dark trees → differential reprod. in pop.
- Adaptations (black colour) benefit from camo provided by black trees → pass on to offspring.
-
Genotype/Allele freq. (mel. variety) cum. ↑
within pop gene pool across gens (Evolution).
Antibiotic Resistance in Bacteria
- Antibiotics: Chemicals produced by microbes that either kill or inhibit growth of bacteria
- Antibiotics are commonly used as treatment for bacterial infections (but not viral infections).
S. aureus:
- S. aureus stim. skin lesions + boils, & pneum.
+ meningitis; treated with antibiotic (methicillin). - Initially, only MSSA strands exist.
- New MRSA strand developed from mutation of antibiotic gene after extensive methicillin use.
- It reproduces & passes on gene to clones.
- MRSA survives & reproduces (passing on gene to clones), whilst MSSA die out (don’t reprod.)
- Gene also transferred to other MSSA in another pop via conjugation (turns into MRSA)
- Results in cumulative change in allele freq. in S. aureus strands → evolution.
Speciation
- Speciation: Gradual divergence of 2 related pops into diff. species due to geographical separation, which → adaptations.
- Degree of divergence depends on extent of geographical separation & amount of time since separation occurred:
- Pops separated recently & are close show less variation (less divergence).
- Pops separated long ago & are far show
more variation (more divergence).
- As genetic divergence between related pops ↑, their genetic compat. consequently ↓.
- When 2 pops diverge to point where no longer interbreed & produce fertile, viable offspring
= separate species (speciation). - Endemic Species: Only found in certain geographical area.
Isolation Barriers

Types of Speciation
Allopatric Speciation: Occurs in diff geographical area. Requires a physical barrier to gene flow.
Sympatric Speciation: Occurs in same geographical area. Requires behavioural or temporal barriers to gene flow.
Both lead to genetically isolated pops; up to the point they can no longer interbreed → new species forms;
Pace of Speciation
- Evolution via speciation may occur via either:
-
Phyletic Gradualism: Speciation generally occurs uniformly, via steady & gradual transformation of whole lineages.
- Supported by fossil record of species
with many intermediate forms connecting ancestral species to modern equivalent.
- Supported by fossil record of species
-
Punctuated Equilib: Speciation is a periodic process that occurs abruptly & rapidly after long periods of stability.
- In this view, speciation is seen as a periodic process (big changes occur suddenly, followed by long periods of no change)
- Supported by general lack of transitional fossils for most species
- But absences could also be due to
unusual & specific events in fossilisation.
Darwin’s Finches
- Charles Darwin’s Theory of Natural Selection used evidence from observing finches in Daphne Major island (in Galapagos).
- Adaptive Radiation: Growing discrepancy of structures from same ancestral line as they perform diff. functions in diff. species due to diff. env. pressures.
- “Darwin’s finches” are a bird species endemic to Daphne Major island in Galapagos.
- Darwin’s finches show adapt. rad. & show beak size & shape (inherited var/adaptation) dep. on
size of seeds available (env. press):-
Drought in 1977: ↓ small seeds / ↑ large
seeds → adv to have larger beak → ↑ in large beak finches / ↓ in small beak sizes. -
Floods in 1983: ↑ small seeds / ↓ large
seeds → adv. to have small beak → ↓ in
large beak finches / ↑ in small beak sizes.
-
Drought in 1977: ↓ small seeds / ↑ large
Clade Reclassification
- Cladograms have shown that Morphology-based class. doesn’t always correspond with evolutionary origins of group of species.
-
3 Outcomes of reclassification:
- Likely closer to truly natural class. (if new classifications based on cladistics); so predictive value = higher.
- Unnoticed similarities between groups & diff. between species previously assumed to be similar revealed.
- Time-consuming & potentially disruptive for biologists.
-
Reclassification of figwort family:
- Until recently, figworts were one of largest family of angiosperms, but problematic as many of figwort plants too diff in structure to function as a meaningful grouping.
- Taxonomists compared chloroplast gene base seq in figworts & other sim genera & reclassified fam into diff clades.
- Now
Binomial System
- Binomial System: Formal system by which all living species are classified (taxonomy)
- Periodically assessed + updated at series across international congresses.
- Good because:
- International system;
- Names agreed at congresses.
- All scientists use the same names for species, which prevents miscomm due to language diff.
- First name is the genus name and shows which other species are closely related, so traits can be predicted for new species
Genus and Taxonomy
- Drunk Katy Perry Comes Over For Great Sex.
- Domain, Kingdom, Phylum, Class, Order, Family, Genus, Species
- All living orgs classified into 3 domains
- Eukarya: Eukaryotes that contain memb-bound nucleus.
- Eubacteria: Prokaryotes lacking nucleus & consist of common pathogenic forms (e.g. E. coli, S. aureus, etc.)
- Archaea: Prokaryotes lacking nucleus consist of extremophiles (e.g. methanogens, thermophiles, etc.)
- Genus: A group of species that share certain characteristics.

Artificial and Natural Classification
- Artificial Classification: Grouping species together solely based on physical characteristics.
- Natural Classification: Grouping species together based on
-
Artificial Disadv is that structures may appear different in similar orgs and sim in diff orgs:
- Div-Evo: Growing discrepancy of structures from same ancestral line as they perform different functions in diff species.
- Conv-Evo: Growing assimilation of structures with different ancestral line as they perform same/sim functions in diff species.
-
Dichotomous Keys: Consist of pairs of choices;
whereby each choice in pair leads to another pair of choices or gives the identification.- Requires a good specimen for reliable ID.
- Key should only use clear/reliable traits.
Plant Phyla

Animal Phyla

Chordata Classes

Clades
- Cladograms: Tree-like diagrams; used to show evolutionary history.
- Clade: Group of organisms consisting of all descendants from a common ancestral org.
- Clade members share inherited characteristics; due to their shared evolutionary history.
- Nodes represent represent common ancestors
as well as when speciation occurred, thus represents sequence in which groups diverged - Clades based on AA seq differences between organisms; # of diffs ∝ how close orgs are.
- Cladograms have led to re-classification of some groups (e.g. figworts).
- Cladistics allow predictions to be made; and estimations of how long ago groups diverged, due to “molecular clock”:
- Mutation rates generally constant, so used as a “molecular clock” to predict when speciation occurred (indicated by branch length).
- Molecular clock limited by:
- Diff genes/proteins change at diff rates
- Over long time, earlier changes may be reversed by later changes → confounding accuracy of predictions.
- Rate of change for particular gene may differ between different groups of orgs
Speciation by Polyploidy
- Speciation: Formation of new species;
- Polyploidy: Form of sympatric speciation due to chrom pairs failing to separate during meiosis; or cell failing to divide in cytokinesis
- Leads to individuals with multiples of normal chrom number;
- Polyploid indivs can interbreed with one another, but not with diploid indivs as it would lead to infertile hybrids → reproductive barrier.
- Common in plants (e.g. Allium genus);


