Clinical Genetics: Chromosomal abnormalities I Flashcards
What form of DNA do chromosomes usually exist as?
- Chromosomes usually exists as chromatin
- DNA double helix wound around an octamer of histone proteins
- Octamer of histones form nucleosome
- Nucleosomes packaged together with scaffolding proteins to form chromatin

What is the difference between euchromatin and heterochromatin?
-
Euchromatin
- Uncondensed, dispersed through nucleus
- Allows gene expression
-
Heterochromatin
- Highly condensed, genes not expressed
DNA is usually loosely packaged within the chromosome. When is this not the case?
- Not the case during cell division when DNA is complexed with various proteins and undergoes several levels of compaction through coiling and supercoiling
What are homologous chromosomes?
- Homologous chromosomes are a pair of identical chromosomes, same length, genes and centromere position.
- One of the pair of chromosomes is inherited from your mother and the other inherited from your father

What is a gene locus?
- A gene locus is the location of a particular gene on a chromosome

What is an allele?
- An allele is an alternate form of a gene
- At each gene locus an individual has 2 alleles, one from each homologous chromosome

Why are chromosomes sometimes shown with a single chromatid?

- Chromosomes with single chromatid show how chromosomes look during interphase - after cell division
Why are chromosomes sometimes shown with two sister chromatids?
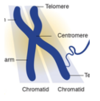
- Chromosomes with 2 sister chromatids show how chromosomes look after S phase where DNA is duplicated in anticipation of cell division
Briefly describe the stages of the cell cycle
- G1 - Cellular contents, except chromosomes are duplicated, Cell makes proteins needed for DNA replication
- S phase - Chromosomes are replicated so that each chromosome now consists of two sister, identical chromatids
- G2 - Synthesis of proteins especially microtubules
- Mitosis - Cell divison

How many pairs of chromosomes do humnas have?
- 23 pairs of chromosomes
- 22 pairs autosomes, 1 pair sex chromosomes XX or XY
What are the 3 different types of chromosome? State which chromosomes within the human genome belong to each type of chromosome
-
Metacentric - p & q arms even length
- 1-3, 16-18
-
Submetacentric - p arm shorter than q
- 4-12, 19-20, X
-
Acrocentric - Long q, small p; p contains no unique DNA
- 13-15, 21-22, Y

What are the different types of chromosomal changes and how can each type be detected?
-
Numerical changes - Can be detected through:
- Traditional karyotyping
- FISH
- QF-PCR (Quantitative fluoresence PCR)
- NGS
-
Structural changes - Can be detected through:
- Traditional karyotyping
- FISH
What is meant by the term “Haploid”?
- One set of chromosomes (n=23) as in a normal gamete
What is meant by the term “Diploid”?
- Cell contains two sets of chromosomes (2n=46; normal in human)
What is meant by the term “Polyploid”?
- Any chromosome number which is an exact multiple of the haploid number e.g. 4n=92
What is meant by the term “Aneuploid”?
- Any chromosome number which is not an exact multiple of haploid number - due to extra or missing chromosome(s) e.g. 2n+1=47
What are the different types of numerical chromosomal abnormalities?
- Trisomy - Type of aneuploidy in which there are three instances of a particular chromosome, instead of the normal two
- Monosomy - Type of aneploidy in which there is only one instance of a particular chromosome
-
Mosaicism - When a person has 2 or more populations of cells with a different number of chromosomes
- E.g. person may have a population of haploid cells (46 chromosomes) and another population of cells with aneuploidy
Give a brief overview of Meiosis
- DNA is replicated so each chromosome has 2 sister chromatids
- Recombination occurs between homologous chromosomes
- Meiosis I: homologous chromosomes line up next to each other at the equator of the cell; get attached to the mitotic spindle and then get separated to opposite spindle poles
- Now each of the 2 cells has 23 pairs of chromosomes (diploid)
- Meiosis II: sister chromatids line up at the equator of the cell and get attached to the mitotic spindle and get separated to opposite spindle poles
- Now each of the 4 daughter cells only has 23 chromosomes (haploid)

Give a brief overview of Mitosis
- Prophase: Chromosomes condense, centrosomes move to opposite poles, mitotic spindle forms
- Prometaphase: Breakdown of nuclear envelope, chromosomes attach to mitotic spindle
- Metaphase: Centrosomes are at opposite poles, Homologous chromosomes line up one behind the other at the equator of the mitotic spindle and get attached to mitotic spindle
- Anaphase: Sister chromatids separated to opposite spindle poles
-
Telophase: chromosomes decondense, nuclear envelope reforms
- Now each of the 2 cells has 23 pairs of chromosomes (diploid)

What is it called when chromosomes/chromatids are pulled to opposite ends of the cell during anaphase?
- Disjunction
How does aneuploidy arise?
- Primary mechanism by which aneuploidy arises is non-disjunction - when homologous chromosomes DON’T separate from one another

What happens if non-disjunction occurs during meiosis I?
- If non-disjunction occur during meiosis I both copies of a pair of homologous chromosomes will end up in one daughter cell while the other daughter cell doesn’t get any
- Meiosis II will occur as normal so daughter gametes formed from cell that got both pairs of homologous chromosomes will end up with 2 copies of that particular chromosome (disomic)
- Daughter gametes formed from cell that didn’t get either pair of the homologous chromosomes don’t have any copies of that particular chromosomes (nullisomic)

What happens if non-disjunction occurs during meiosis II?
- If non-disjunction occurs during meiosis II then both sister chromatids of a particular chromosome end up in one daugther gamete while the other daughter gamete doesn’t get any
- This means one daughter gamete is disomic with respect to that chromosome while the other is nullisomic

Give some examples of the most common autosomal trisomies
- Trisomy 21
- Trisomy 18 (Edward’s syndrome)
- Trisomy 13 (Patau Syndrome)