Chromatin and Transcription Initiation and elongation Flashcards
(38 cards)
Histone Acetylation
- Importance of post-translational modifications of the histone tails.
- Changes in charge (Acetyl group is negatively charged) will promote nucleosome dissociation. It also may change the potential protein partner that could bind to the histone tails.
- The acetylation is reversible (HAT vs. HDAC). At least 10 different HDACs have been identified. HAT exists in two forms HAT-A (nuclear) and HAT-B (cytoplasmic)
N-terminal tail
histone-fold domain
Lysine modified by addition of acetyl group (HAT) or removal (HDAC)
Histone Acetylation
HAT can form a complex with an activator
HDAC (1 to 12) can form a complex with a repressor
Coactivator (HAT)+ Activator + acetyl group =Acetylated histones-active chromatin
Co-repressor(HDAC) + repressor=deacetu;ate histones - inactive chromatin
Hat is a co-activator (often needs to be brought in contact with histone tails). HDAC is a co-repressor requiring a partner to find histone tails
histone methylation and phosphorylation
Active chromatin - H3 N-terminal tail
Inactive chromatin
the histone N-termini are the target of the modification
Multiple methyl groups can be present (tri-methylation of Lys 4,9 or histone H3, Lys 20 on H4, play a role in regulation
Other histon modifications: Phosphorylation and methylation. Different charges and conformations will dictate the function
Summary of location and type of histone tail modifications

What is the role of histone in post-translational modifications
- Repression/silencing
- Inprinting (silent allele)
- Activation
Sumo: Small Ubiquitin-like modifier 18% identity with Ub, similar 3D structure
TSS: transcription start site
Nucleosome remodeling factors
Binding of transcriptional activatio and chromatin remodeling faction
Chromatin remodeling factor + activator
Nucleosome displacement
Binding of general transcription factors and RNA polymerase
What is the role of nucleosome remodeling complexes
sliding, changes in DNA path partial dissociation
ATP dependence (through SWI2 or ISWI sub-unit)
a. translocation-dependent remodeling = SWI/SNF and ATP is converted to ADP considered Nucleosome sliding
b. remodeling of DNA contacts
ATP to ADP SWI/SNF Changes in DNA path
c. remodeling of octamer structure
Loss of H2A-H2B dimer
Swi/Snf, NURF, CHRAC, NURD
All complexes are ATP-dependent
Epigenetic inheritance of histone modifications
- Parental chromatin
- DNA replication
- Parental nucleosomes distributed to progeny strands
- Incorporation of new nucleosomes
- Parental modified histones direct modification of newly incorporated histones
Regulation of transcription by siRNAs
- siRNA-association with RITS unwinding of siRNA
- Pairing with mRNA transcript at target gene
- mRNA + siRNA + RITS
- methylation of H3 lysine-9 heterochromatin formation
- transcription repressed
RITS: RNA-induced initiation and transcriptional Gene silencing
Chromatin Structure and Enhancers
Role of nucleosomes as a structural element that will redirect the DNA path and allow long distance interactions
here is an example of how the enhancer region can be brought to close proximity with the promoter region
the protein binding the enhancer region can now interact with TF or mediators
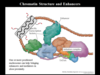
Insulator
Euchromatin
Insulator
Heterochromatin
Role: Prevent spreading of heterochromatin to transcriptionally activ regions
chromatin domain are separated by DNA sequences called insulators
Trans acting factors are maintaining the structure of the insulator region in the proper configuration avoiding loss of cohesion
the euchromatin is organized in large loops that can undergo torsonial stress (changes in topology) that can be important for chromatin remodeling
Chicken B-globin gene cluster
Proteins such as CTCF (CCCTC-binding factor, a zinc-finger protein) can bind to the 5’ hyper=sensitive region HS4 preventing “chromatin invasion”
Concept of Boundary elements where a specific region prevents heterochromatin from invading the region that needs to be transcribed
Boundary elements are cis elements (sequences) that can be bound by specific DNA-binding proteins (example for beta globin: CTCF is a cCCTC-binding factor (zinc finger protein)
DNase insenstive = Low histone acetylation
DNase senstive = high histone acetylation
regulation of transcription in bacteria
Metabolism of lactose
Lac Operon: lactose is a precursor of glucose
regulation of transcription based on need for metabolism intermediates
Glucose requred will necessitate the lactose to be broken into glactose plus glucose
then enzyme required is the B-galactosidase
Lac Operon
- In the absence of lactose with a repressor = no lac nRNA
I, p, repressor, z, y, a
- Presence of lactose
I, p, o, z, y, z
z=galactosidase, y=permease, a=transacetylase
Transition for actively binding repressor vs inactiv binding
Positive Control of the lac Operon by glucose
POSITIVE CONTROL:
- The production of B-Galactosidase is controlled at the level of the Promoter-Operator region (operon).
- The lac z gene codes for ß-galactosidase
- Transduction of the signal for LOW GLUCOSE is transduced to an enzyme named ADENYLATE CYCLASE which role is to hydrolyze ATP to cAMP plus P~Pi (pyrophosphate).
- cAMP will form a complex with the protein CAP (not to be confused with the cap structure at the end of mRNA 7 methyl guanosine).
- The complex will bind DNA and the bacterial RNA polymerase at the Promoter-operator site.
CAP: Catabolic Activator Portein
P: promoter region
O: operator
Z: B-galactosidase

Negative control of the lac Operon
- NEGATIVE CONTROL: Repression.
- In presence of Lactose, no need for
- The expression of the structural genes is not only influenced by the presence or absence of the inducer, it is also controlled by a specific regulatory gene. The regulatory gene may be next to or far from the genes that are being regulated.
- The regulatory gene codes for a specific protein product called a REPRESSOR.
- The repressor acts by binding to a specific region of the DNA called the operator which is adjacent to the structural genes being regulated.
- The structural genes together with the operator region and the promoter is called an OPERON.
- However, the binding of the repressor to the operator is prevented by the inducer and the inducer can also remove repressor that has already bound to the operator.
- Thus, in the presence of the inducer the repressor is inactive and does not bind to the operator, resulting in transcription of the structural genes.
- In contrast, in the absence of inducer the repressor is active and binds to the operator, resulting in inhibition of transcription of the structural genes.
- This kind of control is referred to a NEGATIVE CONTROL since the function of the regulatory gene product (repressor) is to turn off transcription of the structural genes.

Regulation of transcription in Eukaryotes
Regulation of transcription in eukaryotes: is more complex
chromatin de-condensation may be a requiremnt for proper expression of genes
Decondensed Chromosome regions in Drosophila also referred to as : Puffs
Identification of Eukaryotic Regulatory Sequences
- The DNA sequences required for proper activation and/or repression have been determined by deletion or addition of DNA within or adjacent to the promoter region(s).
- The regulatory sequence is cloned upstream of a reporter gene, the expression of which can be monitored by colorimetric assay (CAT or Chloramphenicol Acetyl Transferase assay), Northern blot of fluorescence measurements (GFP for example)
- The DNA was cloned into specific vectors (plasmid that can be expressed in Eukaryotic cells).
- Note that the plasmid is not integrated into the chromosome (transient expression will be lost when the plasmid is lost).
By changing the regulatory sequence one can determine the necessary region for proper regulation (ie the proteins that will bind to those sequences)

DNA Methylation
In females the X-chromosome exist in duplicate (XX vs XY)
If both copies of the X-chromosome were active, there would be an imbalance in expression of the genes coded by the X
One of the two X-chromosome is therefore inactivated
The inactivation mechanism involves DNA modification and protein binding to various regions of the X-chromosome
DNA methylation of Cytosine (in CpG dinucleotides) acts as a flag for gene inactivation
Note that specific protein can bind to methylated C (ex. MeCP2 or other MDBs)
Role is to maintain repression of the methylated DNMA regions

Maintenance of Methylation Patterns
Methylated parental DNA
DNA replication
Methylation
Methylated daughter DNAs
How is methylation maintained?
During replication only one strand will have a methyl group at the CpG
The second strand will be methylated by specific DNA methylase enzyme(s)
Maintenance of repressive state: genomic imprinting
the X-chromosome is maintained in a repressed state (phenomenon of gene imprinting)
One copy of the X-chromosome is already methylate (sperm)
After fusion of Egg and Sperm one allele will be active and one will be inactive
If both X-chromosome are un-methylated one of the two alleles will be inactivated
Overview processing eukaryotic mRNAs

modifications of 3’ ends of eukaryotic mRNAs

Overview of splicing: removal of introns in the nucleus
Splicing requires specific sequences to be recognized as intron-exon boundaries, RNA needs to be cleaved and then re-ligated at the exon-exon interface
3 steps:
recognition of intron and exon site
cleavage
joining
Consensus sequence for intron splicing in higer eukaryotes
Consensus sequences at donor and acceptor sites plus presence of the branching point (A).
Donor obeys the GT-AG rule: GT at the 5’end absolutely required) same for AG at the 3’end. The rest of the requirement for sequences is more loose.
Distance bewteen branching point and acceptor site is 20-40 ntdes.