Chapter 12: Synthesis and Processing of Bacterial RNAs Flashcards
(35 cards)
Module 10
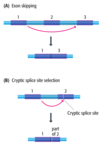
Bacterial mRNA can immediately be translated, however ______ and _____, on the other hand, are first transcribed as pre-RNAs which become functional only after a series of cutting events and chemical modifications
- tRNAs
- rRNAs
Module 10
rRNA processing
- Bacteria synthesize three different rRNAs, called
- the names indicating the sizes of the molecules as measured by _____ _____
- three genes for these rRNAs are linked into a single ______ ______, usually present in multiple copies
- pre-rRNA contains copies of all three rRNAs
- Cutting is needed to release the mature rRNAs. These cuts are made by
- 5S rRNA, 16S rRNA, and 23S rRNA
- sedimentation analysis
- transcription unit
- ribonucleases III, P, and F
Module 10
rRNA processing
steps
- cuts are made by
- ribonucleases III (endonucleases)
- P (ribozyme) and F
- found in all organisms
- cut ends are then trimmed by the exonuclease activities of ribonucleases M16, M23, and M5 (exonucleases) to produce mature rRNAs
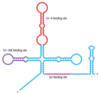
tRNA Processing
Genes for tRNAs are distributed around bacterial genomes, some on their own and some as _____ _____ in multi-tRNA transcription units. In some bacteria, they occur as ______ in the rRNA transcription units, as is the case with E. coli, which has either one or two tRNA genes between _____ _____. All pre-tRNAs are processed in similar though not identical ways
- tandem arrays
- infiltrators
- mRNA genes
Module 10
tRNA Processing
steps
- tRNA sequence within pre-tRNA adopts its cloverleaf structure
- two additional hairpin structures form, one on either side of the tRNA
- ribonuclease E or F cuts just upstream of one of the hairpins, forming a new 3’ end
- Ribonuclease D (exonuclease) trims seven nucleotides from this new 3’ end and pauses, waiting for ribonuclease P
- ribonuclease P makes a cut at the start of the cloverleaf, forming the 5’ end of the mature tRNA
- Ribonuclease D then removes two more nucleotides, creating the 3’ end of the mature molecule
- mature tRNAs must end with the trinucleotide 5¢–CCA–3¢.
- in some pre-tRNAs this sequence is absent, or is removed by the processing ribonucleases
- removal occurs when 3’ ends are created by an endonuclease called ribonuclease Z
- makes a cut adjacent to the first base pair in the tRNA cloverleaf removing the region containing terminal CCA
- When CCA is absent, it’s added by one or more template-independent RNA polymerases such as tRNA nucleotidyltransferase

one of ribonuclease P’s subunits is made of ____ rather than protein. these _____ subunits are present in several enzymes involved in RNA processing, including ribonuclease _____, which works with ribonuclease P in processing of eukaryotic pre-rRNAs. These hybrid _____-_____ enzymes may be relics of the _____ world
- RNA
- RNA
- MRP
- protein–RNA
- RNA
Module 10
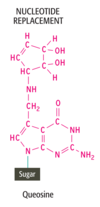
The final type of processing that occurs with pre-RNAs is the chemical modification of ______ within the transcript. This occurs with _____ and ______. A broad spectrum of chemical changes has been identified with different pre-RNAs: over 50 different modifications are known in total. Most of these are carried out directly on an existing nucleotide within the transcript but two modified nucleotides, ______ and ______, are put in place by cutting out an entire nucleotide and replacing it with the modified version
- nucleotides
- prerRNAs
- pre-tRNAs
- queosine
- wyosine
Module 10
Many of these unusual nucleotides were first identified in tRNAs, where one in ten nucleotides becomes altered. Reasons why these modifications may occur are
- may mediate the recognition of individual tRNAs by the enzymes that attach amino acids to these molecules
- increases the range of the interactions that can occur between tRNAs and codons during translation
- enabling a single tRNA to recognize more than one codon
Ribosomal RNAs are modified in two ways:
- addition of methyl groups to, mainly, the 2’–OH group on nucleotide sugars, and by conversion of uridine to pseudouridine
- occurs at the same position on all copies of an rRNA
- same in different species
- bacterial rRNAs are less heavily modified than eukaryotic ones
- function behind modifications have not been identified
- most occur within those parts of rRNAs thought to be most critical to the activity of these molecules
- Often two or more nucleotides in the same region are modified at once
Module 10
RNA degradation is equally important, especially with regard to ______ whose presence or absence in the cell determines which proteins will be ______. Degradation of specific mRNAs could be a powerful way of ______ genome expression. rate of degradation of an mRNA can be estimated by determining its ______ in the cell. There are considerable variations between/within organisms. Bacterial mRNAs are generally turned over very rapidly, their half-lives rarely being longer than a ______ minutes while Eukaryotic mRNAs are longer lived, with half-lives of, on average, _______ minutes for yeast and several ______ for mammals
- mRNAs
- synthesized
- regulating
- halflife
- few
- 10–20
- hours
Module 10
Bacterial mRNAs are degraded in the ______ direction
3’→5’
Module 10
RNA-degrading enzymes that are thought to be involved in mRNA degradation
- RNase E and RNase III
- endonucleases that make internal cuts in RNA molecules.
- RNase II
- exonuclease that removes nucleotides in the 3’→5’ direction
- Polynucleotide phosphorylase (PNPase)
- exonuclease that removes nucleotides in the 3’→5’ direction
- requires inorganic phosphate as a cosubstrate
Module 10
RNA-degrading process
- most mRNAs have a hairpin structure near the 3’ end
- hairpin that induced termination of transcription
- The termination hairpin blocks the exonuclease activities of RNase II and PNPase
- termination hairpin is removed by endonuclease action of RNase E and/or RNase III
- removal exposes a new end for RNase II and PNPase to attach and destroy the functional activity of the mRNA
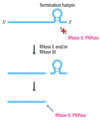
degradosome
- contains RNase E and PNPase
- include an RNA helicase
- might be involved in both rRNA and mRNA degradation.
- not clear and a few researchers are sceptical about its actual existence,
Module 10
Synthesis and Processing of Eukaryotic RNA
promoter clearance
transition from the preinitiation complex to a complex that has begun to synthesize RNA
Module 10
Synthesis and Processing of Eukaryotic RNA
promoter escape
when polymerase moves away from the promoter region and becomes committed to making a transcript
Synthesis and Processing of Eukaryotic RNA
The opposing effects of negative and positive _____ ______ influence the ability of the polymerase to begin productive RNA synthesis, and if the ______ factors predominate then transcription halts before the polymerase has moved more than ______ nucleotides from the initiation point. Promoter escape could therefore be an important _____ _____
- elongation factors
- negative
- 30
- control point
Module 10
Synthesis and Processing of Eukaryotic RNA
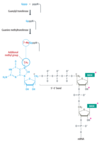
Capping of RNA polymerase II transcripts
- occurs immediately after initiation
- Successful promoter escape could be linked with capping
- completed before the transcript reaches 30 nucleotides
- an extra guanosine is added to the extreme 5’ end of the RNA
- involves a reaction between the 5’ triphosphate of the terminal nucleotide and the triphosphate of a GTP nucleotide
- the outermost phosphate is removed, as are the β and γ phosphates of the GTP, resulting in a 5’–5’ bond
- carried out by the enzyme guanylyl transferase
- conversion of new terminal guanosine into 7-methylguanosine by attachment of a methyl group to nitrogen number 7 of the purine ring
- catalyzed by guanine methyltransferase
- called a type 0 cap
- commonest form in yeast
- In higher eukaryotes, additional modifications occur
- A second methylation replaces the hydrogen of the 2’–OH group of what is now the second nucleotide in the transcript. This results in a type 1 cap.
- If this second nucleotide is an adenosine, then the amino group attached to carbon number 6 of the purine ring might also be methylated.
- Another 2’–OH methylation might occur at the third nucleotide position, resulting in a type 2 cap.
- two capping enzymes make attachments with the CTD
- possible that they are intrinsic components of the RNA polymerase II complex during promoter clearance
Module 10
Synthesis and Processing of Eukaryotic RNA
All RNAs synthesized by RNA polymerase II are capped in one way or another. This means that as well as mRNAs, the ______ that are transcribed by this enzyme are also capped. The cap may be important for
- snRNAs
- Promoter escape by RNAP II
- Transport to cytoplasm
- Initiation of translation
- Stability of mRNA
Elongation of eukaryotic mRNAs
bacterial genes can be transcribed in a matter of minutes In contrast, RNA polymerase II can take hours to transcribe a single gene because the presence of multiple ______ in many eukaryotic genes means that considerable lengths of DNA must be copied. the extreme length places demands on the stability of the _____ ______.
- introns
- transcription complex
Elongation of eukaryotic mRNAs
In the nucleus, pausing and stopping pausing of RNA polymerase II can occur when the enzyme transcribes through a region where intrastrand base pairs (e.g., a hairpin loop) can form. This pausing/stopping is reduced by ______ ______ which are …. ______ are currently known
- elongation factors
- proteins that associate with the polymerase after it has cleared the promoter and left behind the transcription factors involved in initiation. Thirteen elongation factors
- 13
Elongation of eukaryotic mRNAs
eukaryotic nuclear polymerases, has to negotiate the ______ that are attached to the template DNA that is being transcribed. The solution to this problem is probably provided by _____ ______ that are able to modify the_____ _____ in some way.
- nucleosomes
- elongation factors
- chromatin structure
Module 10
Termination mRNAs - polyadenylation
Most eukaryotic mRNAs have a series of up to ____ adenosines at their_____ ends. These As are not specified by the _____ and are added to the transcript by a template-independent RNA polymerase called _____ _____. This polymerase does not act at the extreme 3’ end of the transcript, but at an _____ _____ which is cleaved to create a new 3’ end to which the poly(A) tail is added. an inherent part of the mechanism for termination of transcription by RNA polymerase II.
- 250
- 3’
- DNA
- poly(A) polymerase
- internal site
Module 10
Termination mRNAs - polyadenylation
the process
- directed by a signal sequence in the mRNA 5’–AAUAAA–3’
- located between 10 and 30 nucleotides upstream of the polyadenylation site
- often immediately after the dinucleotide 5’–CA–3’
- followed 10–20 nucleotides later by a GU-rich region
- Both poly(A) signal sequence and the GU-rich region are binding sites for multisubunit protein complexes
- cleavage and polyadenylation specificity factor (CPSF)
- cleavage stimulation factor (CstF)
- Poly(A) polymerase, PADP, and at least two other protein factors associate with bound CPSF and CstF
- one of them is polyadenylatebinding protein (PADP)
- helps polymerase to add the adenosines
- possibly influences the length of the poly(A) tail
- may play a role in maintenance of the tail after synthesis
- one of them is polyadenylatebinding protein (PADP)
Steps
- CPSF interacts with TFIID and is recruited into the polymerase complex during the transcription initiation stage
- Rides along twith RNA polymerase II, and binds to the poly(A) signal sequence as soon as it is transcribed, initiating the polyadenylation reaction
- Both CPSF and CstF form contacts with the CTD of the polymerase
- these contacts may change when the poly(A) signal sequence is located
- this change alters the properties of the elongation complex so that termination becomes favored over continued RNA synthesis
- transcription stops soon after the poly(A) signal sequence has been transcribed