Cellular processes, control (2) + (5) Flashcards
(149 cards)
where does glycolysis occur
the cytoplasm
briefly describe the difference between the presence of oxygen for cellular respiration in plants/fungi and animals
The presence of oxygen will allow the glucose (now as two molecules of pyruvate after glycolysis) to be fully oxidised.
Oxygen acts as the final electron acceptor in the ETC, allowing NAD and FAD to be oxidised. Therefore the Krebs cycle and Link reaction and ox. phos. can continue to convert pyruvate.
In the absence of oxygen, pyruvic acid is broken down into ethanal and carbon dioxide, and not fully oxidised. Pyruvic acid is converted to lactic acid and carbon dioxide is released. Glucose is not fully oxidised.

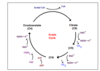
where does the Krebs cycle occur
in the mitochondrial matrix

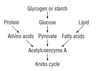
- outline what pyruvate is converted to in Links*
- and what happens to it in the Krebs cycle*
Pyruvate (2C) is converted to Acetyl CoA (2C).
Acetyl CoA binds to 4C intermediate, producing citric acid (6C).
It is oxidised and decarboxylated to produce a 5C intermediate.
That is oxidised and decarboxylated to produce a 4C intermediate (which binds to 2C pyruvate derivative, continuing the cycle).

how much ATP is produced in aerobic respiration
there is a net gain of 38 ATP molecules from one molecule of glucose
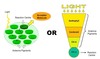
how do animals and plants make glucose
Plants are photoautotrophs/chemoautotrophs- absorb light energy and use CO2 and water to produce glucose.
Animals consume carbohydrates which are hydrolysed into glucose or other molecules to be used in respiration.

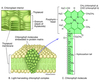
where does photolysis occur
in the granum of the chloroplast
why must DNA produce genetically identical DNA when replicating
old cells/organelles/molecules die and must be replaced, it must be genetically identical so that they have the exact same structure and function as the previous molecules, which is decided by the sequence of nucleotide bases. DNA must also be able to be passed onto offspring.
what is necessary for DNA replication to occur (4)
- the actual DNA to act as an exact template
- a pool of relevant and freely available nucleotides
- a supply of the relevant enzymes to catalyse/speed up the rate of reaction
- ATP to provide energy for these reactions

what happens to the DNA structure during replication
the double helix unzips, uncoils and unwinds so that each strand is exposed for the free nucleotides to bond to
why is the new strand of DNA semi-conservative
50% of the genetic material comes from the original strand, and 50% from free nucleotides and the new strand not previously a part of the original strand
why does nucleic acid sequence determine which protein is produced in protein synthesis
the sequence determines which amino acids are translated, which give a specific sequence which determines how the protein folds and coils, in particular because it determines the R groups, which have different interactions, determining the tertiary structure of the protein
how can the four DNA nucleotides be classified
PYRIMIDINES- cytosine and thymine, single carbon ring structure PURINES- adenine and guanine, double carbon ring structure COMPLEMENTARY BASE PAIRING C and G triple hydrogen bond T and A double hydrogen bond Purines bond with Pyrimidines, which allows a constant distance to always be maintained between the two strands. There is always an equal quantity of A and T, and an equal quantity of C and G.
explain 5’ and 3’ in DNA
the two polynucleotide strands of the double helix are ANTIPARALLEL, running in opposite directions. For one strand the phosphate group end (5’) is at the top, and for the other the hydroxyl group (3’) is at the top. They are bonded together by hydrogen bonds.
what is the function of mRNA and why is it necessary
To transport the genetic material needed to code for proteins at the ribosomes in the cytoplasm from the double-membrane bound nuclear envelope where DNA is protected from the cytoplasm, stored and transcribed. DNA is too large a molecule to leave the nucleus. Therefore its genetic code is transcribed onto mRNA. mRNA corresponds to a single gene, compared to DNA, which is an entire chromosome long.
differences and similaritiesbetween DNA and RNA
D1. SUGAR- deoxyribose/ribose, 4 O atoms, 5 O atoms D2. thymine is replaced by URACIL (still pyrimidine, double bonds with A) D3. RNA polymers are small enough to leave the nucleus and travel to ribosomes D4. RNA is involved in protein synthesis S1. complementary base pairing rules- number of H bonds and between what S2. nucleotides form polynucleotides in the same manner- phosphodiester bonds between Pi on C5 and OH on C3
why does DNA need to be replicated
for new cells needed for growth or repairof tissues
what is DNA replication
the process of copying and duplicating a DNA molecule in a semiconservative way, i.e. the copy contains one of the original strands paired with a newly synthesized strand that is complementary in terms of AT and GC base pairing
outline the 3 stages of DNA replication
- the DNA double helix structure must unwind and 2. the H bonds holding the two strands together must be hydrolysed by DNA HELICASE, which travels along the sugar-phosphate backbone, leaving the nucleotides exposed 2. free nucleotides H bond to the exposed nucleotides using the complementary base pairing rules, forming H bonds between them 3. the new adjacent nucleotides then form phosphodiester bonds between them, catalysed by DNA POLYMERASE, creating a new polynucleotide
what risk is there during DNA replication for the organism
random and spontaneous mutations may occur, where complementary base pairing is not followed and leads to an incorrect sequence of bases
in what way is the genetic code carried before translation
in a TRIPLET CODE; each sequence of three bases forms a codon, which codes for a specific amino acid A section of DNA that contains the complete sequence of codons is a gene.
why is the genetic code described as ‘degenerate’
there are 64 possible base triplets/codons (4x4x4) including one start and three stop codons which means that DNA does not overlap There are 20 amino acids, which can be coded for by more than one codon. Therefore the 64 codons are degenerate.
what are the 6 stages in DNA transcription to mRNA
- In the nucleus, RNA polymerase attaches to the DNA molecule. It uncoils and unzips the double helix structure, hydrolysing the hydrogen bonds between the base pairs, starting at the START codon.
- Free RNA nucleotides bond to the exposed bases on the template/antisense strand by their complementary pairs. Adenine bonds with uracil instead of thymine. (C+G)
- Free RNA nucleotides do not bond with the coding/sense strand because it is the strand that needs to be copied in order to translate the correct amino acids to produce specific proteins.
- RNA polymerase catalyses the bonding of nucleotides together by phosphodiester (covalent) bonds in a condensation reaction, which forms an mRNA polynucleotide.
- H bonds form between the two strands and they coil into the double helix structure. The entire process continues until RNA polymerase reaches the STOP codon.
- mRNA detaches from the DNA molecule and leaves the nucleus via a nuclear pore.

name the two strands of DNA that are involved in transcription
- SENSE/ coding strand (5’ to 3’)
- It is transcribed to mRNA but does not bond to free RNA nucleotides because it is the one that needs to be copied.
- ANTISENSE/ template strand (3’ to 5’)
- It is not transcribed to mRNA but bonds to free RNA nucleotides in the nucleus.