CBIO 3.1: Cell Signalling in Cancer Flashcards
Observe the learning outcomes of this session

How is cancer related to cell signalling pathways?
- many common genetic mutations in cancer involve signalling proteins
How many receptor tyrosine kinase (RTK) subfamilies are there?
- there are 20 receptor tyrosine kinase (RTK) subfamilies
- see diagram for the different domains that characterise each subfamily

Describe the RTK subfamily, ErbB
- types
- function
- cancer associations
- 4 highly related types:
- EGFR: also known as ErbB-1 or HER1
- ErbB-2 (HER2)
- ErbB-3 (HER3)
- ErbB-4 (HER4)
- function:
- activated by growth factors of the epidermal growth factor (EGF) family
- they have disulfide bonds that determine bonding specificity
- contain structural motifs:
- immunoglobulin (Ig)-like domains
- heparin-binding sites
- glycosylation sites
- ErbB receptors bind to each other in different combinations
- ErbB2 does not bind ligands itself but acts as a co-receptor for other members of the subfamily
- cancer associations:
- EGFR is found in many different organs and its abnormal expression and/or activation is associated with many types of cancers

Using this diagram, describe how RTKs can induce signalling pathways

- after cross-phosphorylation has occurred, the cytoplasmic tails of RTKs behave as ‘docking’ sites for specific intracellular, cytoplasmic proteins
- figure A shows the several docking sites on the activated EGF receptor where cytosolic signalling proteins can bind
- these proteins have a particular domain, the SH2 domain, that binds to sequences in the cytoplasmic tails of RTKs that contain a phosphorylated tyrosine (figure B)
- RTK tails can contain multiple phosphorylated tyrosines and it is possible for more than one SH2-containing protein to bind to an activated RTK at the same time
- this can promote simultaneous activation of several intracellular signalling pathways
- some signalling proteins, e.g. PLC-γ, bind to the activated RTK directly
- others bind via adaptor proteins such as Shc and Grb2
- in addition to SH2 domains, many signalling proteins contain one or more SH3 domains
- these domains bind to proline-rich sequences on other proteins
- and recruit them to the activated RTK to propagate the growth factor signal
- which causes alterations in gene transcription
- signalling becomes complex as signals are transferred from the membrane to the nucleus, due to possible cross talk that can occur between different intermediates in numerous signalling pathways found within the cell
How do SH2 and SH3 domain-containing proteins interact with activated RTKs?
Give an example
What else are they known as?
Functions
- SH2 and SH3 domain-containing proteins that interact with activated RTKs are known as adaptor proteins
- e.g. Grb2
- these proteins link the activated receptors to other parts of the signalling pathway
- but they themselves don’t have intrinsic signalling abilities
What is Ras and how is it activated?
- Ras is a key signalling molecule in pathways switched on by activation of RTKs
- it is a membrane-bound guanosine triphosphate (GTP) / guanosine diphosphate (GDP) - binding (g) protein
- it acts as a ‘molecular switch’, transforming signals from the cell membrane to the nucleus
- diagram description:
- The Grb2 Adaptor Protein Recruits Sos to the RTK.
- The Grb2 SH2 domain binds to a phosphotyrosine docking site on the RTK.
- The two SH3 domains of Grb2 recruit Sos.
- Sos is a nucleotide exchange factor that swaps GTP for GDP on Ras.

Give examples of some signalling pathways and proteins that are affected by oncogenic mutations
- proteins involved in signalling pathways that are commonly activated in many different cell response
- e.g. the growth factor RTKs:
- EGFR
- specific proteins affected:
- e.g. small GTPases such as Ras
- the serine/threonine kinases Raf and Akt
- cytoplasmic tyrosine kinases Src and Abl
- lipid kinases (phosphoinositide 3-kinases PI3Ks)
- nuclear receptors: e.g. estrogen receptor (ER)
- downstream nuclear targets of signalling pathways
- e.g. transcription factors: c-Myc
- cell cycle effectors: cyclins

What can deletions and other loss-of-function mutations suppress?
Give examples
- they can suppress negative regulators that normally function as tumour suppressors
- e.g. p53: pivotal in controlling cell proliferation and stress responses, such as apoptosis and DNA damage repair
- tumour suppressors can also act as negative regulators of cytoplasmic signalling
- e.g. lipid phosphatase PTEN is a negative regulator of the PI3K-Akt pathway
What is the role of the majority of cell membrane receptors?
- they are capable of receiving and passing on signals from the environment
- some are also able to act as enzymes
- an extracellular, soluble signalling molecule binding to the membrane receptor triggers the receptor’s in-built enzyme activity
How do RTKs form cross-linked dimers?
What does their formation trigger?
- RTKs are bound by and respond to growth factors and other proteins that are present at low concentrations in their surrounding environment
- binding of a signalling molecule causes adjacent/neighbouring RTKs to join with each other, forming cross-linked dimers
- this cross-linking triggers the tyrosine kinase activity in the RTKs through cross phosphorylation
- this means that each RTK within the dimer phosphorylates several tyrosines on the other RTK partner

Describe RTK activation with help from this diagram

- A:
- the two RTKs are shown to be sitting adjacent to each other but with some distance separating them. Each RTK has on its extracellular side a ligand binding site
- A red growth factor/ligand molecule, represented as two circles with tails that crisscross each other, is shown above the RTKs to which it will bind.
- B:
- the two RTKs are shown bound to the growth factor/ligand
- The RTKs have now formed a dimer and are shown to be much closer in position to one another.
- C:
- the RTKs are still bound to the ligand, but now each RTK has some yellow circles labelled with P’s on its outer edge
- This indicates that the activated RTKs have phosphorylated each other on tyrosine residues (known as cross-phosphorylation).
- D:
- the phosphorylated RTKs are bound to different proteins.
- The different proteins, which are shown as dark blue, light blue, and orange shapes, are docked at each phosphorylated site on both of the RTKs
- The pink-coloured ‘rays’ on the outer edge of the intracellular region of each RTK indicate that the signalling proteins are activated.
How is RTK activity kept in check under normal physiological conditions?
- by the low availability of growth factors/ligands
- endocytosis and degradation of activated growth factors and protein tyrosine phosphatases (PTPases)
- PTPases are highly active enzymes that remove the phosphate group (dephosphorylate) phosphorylated tyrosines
What happens to RTK activity in cancer?
- use EGDR as an example
- why this may happen
- gene mutations can result in RTK activation and clustering in the absence of growth factor/ligand
- EGFR:
- activating mutations have been found in the kinase domain, extracellular, transmembrane and juxtamembrane domains
- chromosomal rearrangements can result in the fusion of RTK genes
- e.g. ALK, FGFR and PDGFR
- these genes make proteins containing an oligomerisation domain, resulting in oncogenic proteins whose kinase activity is permanently switched on
- giving cancer cells a growth advantage
How does the binding of a growth factor (e.g. EGF) to an RTK (e.g. EGFR) cause activation of Ras
- Two cytoplasmic proteins, Grb2 and Sos, provide the vital connection
- An SH2 domain in Grb2 binds to a specific phosphotyrosine residue in the activated receptor
- Grb2 also contains two SH3 domains, which bind to and activate Sos.
- Grb2 thus functions as an adapter protein for the EGF receptor, linking it to Sos.
- Sos is a guanine nucleotide exchange protein (GEF), which converts inactive GDP-bound Ras (note that Ras is located in the membrane) to the active GTP-bound form.
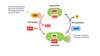
Explain the steps of Ras activation
- A Guanine nucleotide exchange factor (GEF) aids the separation of GDP from Ras.
- GTP then binds spontaneously to Ras, and the GEF detaches, resulting in the active Ras · GTP form.
- Finally, hydrolysis of the bound GTP restores the inactive Ras · GDP form. This process is enhanced a hundred-fold by a GTPase-activating protein (GAP).

What signalling pathways can GTP-bound Ras activate?
- MAPK (MAP kinase) signalling pathway
- phosphoinositide 3-kinase (PI3K)
- Ral-GEF signalling pathways
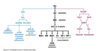
What is the MAP signalling pathway
- the mitogen-activated protein (MAP) kinase cascade is one of the most common intracellular signalling pathways triggered by RTKs
- involves three serine-threonine kinases

Describe the MAP kinase cascade
- the pathway starts with the activation of Ras
- GTP-bound Ras activates Rad, the first serine-threonine kinase in the MAP kinase cascade
- Each of the three kinases in this pathway activates the next by phosphorylating it, as the third is the MAP kinase (in this example Erk1),
- the kinase that phosphorylates MAP kinase is sometimes termed the MAP kinase kinase (MAPKK, in this example MEK)
- the kinase that phosphorylates the MAP kinase kinase (Raf) is termed the MAP kinase kinase kinase or MAPKKK
- As all three kinases in this pathway phosphorylate several different factors, the initial signal can be amplified at each step
- The final enzyme in the pathway then phosphorylates one or more transcription regulators, thus altering gene transcription.

What substances signal via MAPK pathways?
What about different pathways?
- many different growth factors such as:
- nerve growth factor (NGF)
- platelet-derived growth factor (PDGF)
- but it must be noted that not all RTKs use the MAP kinase cascade to transfer signals to the nucleus
- e.g. insulin-like growth factor receptors (IGFRs) act via the PI3K pathway

Describe how Ras activates PI3K and what this stimulates
- Ras activates the enzyme phosphoinositide 3-kinase (PI3K) by binding to the Ras-binding domain (RBD) of PI3K and stimulating its catalytic domain
- Note that PI3K can also be activated directly through RTKs such as PDGFRβ, which contains a docking site for PI3K
- Activated PI3K then phosphorylates phosphatidylinositol 4,5-bisphosphate (PI(4,5)P2 or PIP2) to produce the second messenger, phosphatidylinositol 3,4,5-triphosphate (PI(3,4,5)P3 or PIP3)
- Both PIP2 and PIP3 are phospholipids that reside in the cell membrane.
- The production of PIP3 activates numerous downstream pathways that control cellular functions critical to cancer, such as cell proliferation and apoptosis
- PIP3 does this by binding directly to proteins with PH domains*, such as phosphoinositide-dependent kinase 1 (PDK1) and Akt (also known as protein kinase B or PKB). PIP3 brings PDK1 and Akt to the membrane, where PDK1 activates Akt by phosphorylation
- Activated Akt then inhibits the proapoptotic Bcl-2 family members BAD and BAX, hence promoting cell survival.
- Importantly, the tumour suppressor phosphatase and tensin homologue (PTEN) inhibits the PI3K pathway by dephosphorylating PIP3 to PIP2

What does PI3K signalling promote due to the functions of Akt?

- Akt phosphorylates several other proteins, seen in the table
- causing PI3K signalling to promote cell survival, proliferation and growth

What are the potential side effects of inhibiting the PI3K pathway?
- PI3K activity is important for insulin signalling and metabolism, so attempting to control tumour progression by inhibition of PI3K signalling could lead to decreased insulin sensitivity and diabetes
- PI3K also appears to be required for normal brain function, as decreased PI3K signalling has been associated with schizophrenia
- Drugs that inhibit PI3K signalling for a short period of time in normal tissues might have reversible or treatable side-effects.
- Nevertheless, because tumour cells are often dependent on a hyperactive PI3K pathway, restoring normal levels of PI3K signalling to these cells could be sufficient to slow tumour progression
What percentage of cancers have alterations in the PI3K pathway?
- 30-40% of cancers
- examples in image



