2 - Translation Flashcards
1
Q
Translation
A
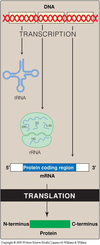
- Translation is the process in which the linear sequence of nucleotides in the mRNA is “translated” into the corresponding protein sequence.
- Transcribed = writing down something in the same language (nucleotide to nucleotide)
- Translated = in different language (nucleotide to amino acids)
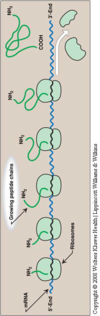
- Amino acids are added on from N-terminus to C-terminus thus C-terminus is the elongating end.
- CO- group from N-terminus and NH- group from the C-terminus combine together into CONH group, thus forming a peptide bonds.
2
Q
The Genetic Code
A
- Written in words three nucleotides long (codons)
- The genetic code consists of 64 codons, each one of which is a triplet of three nucleotides
- Codons have to be 3 nucleotides long in order to encompass all amino acids
- Three codons are stop codons (UAG, UGA, UAA). The start codon (AUG) codes for methionine.
- Sixty one codons code for the standard amino acids; whereas the other 3 are the Stop codons
- Anti-codons read (pair with) the mRNA during translation

3
Q
The Genetic Code (cont.)
A
- Specific: unambiguous, always codes for the same amino acid
-
Universal: almost absolutely conserved throughout evolution
- Some minor differences, e.g. codons found in mitochondria is a little bit different from those found in cell in general.
- Redundant (degenerate): a given amino acid may have more than one triplet coding for it e.g. 6 codons coding for a single amino acid.
-
Nonoverlapping and commaless (the code is read from a fixed starting point as a continuous sequence of bases, taken three at a time):
- ABCDEFGHIJKL
- ABC/DEF/GHI/JKL
4
Q
Mutations
A
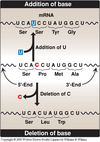
-
Nonsense mutation: The codon containing the changed base may become a termination codon.
- e.g. UCA (serine) –> UAA (Termination codon)
-
Missense mutation: The codon containing the changed base may code for a different amino acid.
- e.g. UCA (serine) –> CCA (proline)
-
Silent mutation: mutation in which there is no change in the encoded amino acid.
- e.g. UCA (serine) –> UCU (serine)
- Nonsense and Missense mutations are very devastating whereas silent mutation has no apparent effect.
5
Q
Mutations (cont.)
A
- There are a number of human neurological diseases caused by triplet expansions.
- There are a number of different mechanisms by which the expansion causes disease.
- Some of these expansions involve the trinucleotide CAG, which are translated into a series of glutamine residues (poly Q). Huntington disease is a polyQ disease.
- Amplification of the CAG codon leads to the insertion of many extra glutamine residues in the huntingtin protein, causing the neurodegenerative disorder, Huntington disease
- Huntington disease is neurodegenerative; dominant disorder; associated with being old
- Huntington is passed down the gene and only show symptoms later down the road when a person is old (not early).
- Non-polyglutamine diseases (expansion of trinucleotides other than CAG) include Fragile X syndrome, myotonic dystrophy, etc.
- Trinucleotide repeat disorders often display genetic anticipation, a process characterized by the addition of further repeats with each successive generation; get worse and worse and eventually reach a point where symptoms are visible.

6
Q
Frame shift mutations
A
- Most of the cases of cystic fibrosis are caused by deletion of a single codon; maintain the reading frame since three nucleotides are lost
- Hemoglobin Gun Hill is caused by the deletion of FIVE codons i.e. 5 amino acids (according to google search).
- Frame shift mutations are responsible for a number of cases of Duchenne muscular dystrophy.
- Duchenne = serious
- Becker’s = less serious form
7
Q
tRNA
A
- The anticodon on the tRNA hydrogen binds to the codon on the mRNA. This binding is antiparallel.
- A charged tRNA molecule has the specific amino acid bound at its 3’end; ultimately deciding which AA is bound [Note: When a tRNA has a covalently attached
amino acid, it is said to be charged; when it does not, it is said to be uncharged.] - However, if, for example, a tRNA that is supposed to carry Methionine is mischarged to say, Cysteine, the anticodon of the mischarged tRNA will still work properly and thus place Cysteine in an AA chain where Methionine should’ve been placed instead.
- In other words, once an amino acid is attached to a tRNA molecule, only the anticodon of that tRNA determines the specificity of incorporation.

8
Q
tRNA (cont.)
A
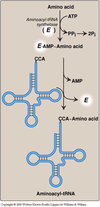
- Attachment of an amino acid to its corresponding tRNA by aminoacyl-tRNA synthetase (E in the figure)
- Aminoacyl-tRNA synthetases catalyze a two-step reaction that results in the covalent attachment of the
carboxyl group of an amino acid to the 3’-end of its corresponding (cognate) tRNA. - The attachment of the amino acid to the tRNA is catalyzed by aminoacyl-tRNA synthetase. Each aminoacyl–tRNA synthetase recognizes both a specific amino acid and the tRNA specific for that individual amino acid. A poor codon–anticodon match will result in the dissociation of the tRNA from the A site of the ribosome –> “proofreading” function.
- The overall reaction requires adenosine triphosphate
(ATP),which is cleaved to adenosine monophosphate (AMP) and inorganic pyrophosphate (PPi). - The experimental modification of the charged amino acid to one different than the one corresponding to the anticodon has demonstrated that the mischarged amino acid is incorporated into the growing polypeptide chain in response to the tRNA’s contact with the codon; this modification, however, is NOT self-corrected.
9
Q
Composition of a ribosome
A
- The rRNA molecules are coordinately expressed
- The ribosome has 3 binding sites for the tRNA molecules, the E, P, and A. These sites extend over both the large and the small subunits (all inside ribosomes).
- E = Empty
- P = Peptidyl
- A = Amino-acyl
- In eukaryotes, the ribosomes can either be free in the cytosol or be associated with the endoplasmic reticulum (rough ER). The ribosomes in the cytosol synthesize proteins that will be localized in the cytosol, nucleus, mitochondria, or peroxisomes thus for its own use.
- The RER-associated ribosomes are responsible for synthesizing proteins that are to be exported from
the cell, as well as those that are destined to become incorporated into plasma, endoplasmic reticulum, or Golgi membranes, or imported into lysosomes - There are specific factors necessary for initiation, elongation, and release. There are more eukaryotic factors than prokaryotic factors for initiation and elongation. There is only one eukaryotic release factor (and two prokaryotic factors).
- Eukaryotes have more initiaion & elongation factors
- Prokaryotes have more release factor
- The cleavage of four high energy bonds, supplied by the hydrolysis of both ATP and GTP, is needed for the incorporation of one amino acid into the polypeptide chain i.e. require substantial amount of energy.

10
Q
Codon Recognition by tRNA
A
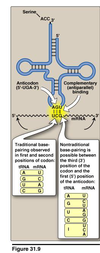
- Recognition of a particular codon in an mRNA sequence is accomplished by the anticodon sequence of the tRNA
- Antiparallel binding between codon and anticodon
- tRNAs can recognize more than one codon for a specific amino acid (the “last” base of the codon “wobbles”)
- Wobble means that there doesn’t have to be tRNA’s for every single possible codons; fewer tRNA (about half of as many codons) are all that’s really needed since a single tRNA can bind to multiple codons
- Traditional base-pairing observed in first and second positions of codon
- Nontraditional base-pairing is possible between the third (3’) position of the codon and first (5’) position of the anticodon
- Many cells have only about 30-some species of tRNA. It is wobble that allows 30-some tRNA molecules to bind 61 different codons.
- The result of wobble is that there need NOT be 61 tRNA species to read the 61 codons that code for
amino acids.
11
Q
Differences between Eukaryotic and Prokaryotic translation
A
- In prokaryotes transcription and translation are closely linked; no nucleus like eukaryotic cells do.
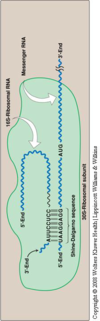
- Prokaryotic mRNA have a Shine-Dalgarno sequence 6-10 bases upstream of the initiating start codon, which is near its 5’-end. This sequence allows the binding of the 16S ribosomal RNA; an example of consensus sequence; rRNA will bind here and orient for the initiation of translation
- The 5’ end of the mRNA and the 3’-end of the 16S rRNA can form complementary base pairs,
- Prokaryotic mRNA is often polycistronic (one long mRNA that codes for many proteins that eventually get cleaved and yield multiple proteins), while eukaryotic mRNA is NOT.
12
Q
Generation of the initiator N-formyl-methionyl-tRNA
A
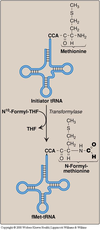
- The start codon (AUG) is recognized by a special initiator tRNA. In prokaryotics (and also in mitochondria) the initiator tRNA carries an N-formylated methionine at the 3’CCA end.
- Peptidyl transferase is a ribozyme (catalytically active RNA molecule).
- N-formyl-methionine is only found in prokaryotes
13
Q
Formation of peptide bond (Prokaryotic translation)
A
- Initiation factors aid in the formation of the 30S initiation complex; at this point, the charged
initiator tRNA (with fMet) is already situated in the P site on the small subunit. - GTP is cleaved and initiation factors are released when the 50S subunit arrives to form the 70S initiation complex.
- Elongation factors direct the binding of the appropriate tRNA to the codon in the empty A-site.
- Peptidyltransferase, an activity of the rRNA of the 50S ribosomal subunit, transfers the amino acid (or peptide chain) from the P-site onto the amino acid at the A-site, and catalyzes peptide bond formation.
- The ribosome moves a distance of three nucleotides along the mRNA in the 5’–>3’ direction. What was in the P site is now in E; what was in the A site is now in P, and the A site is empty.
- Step 3, 4, and 5 are repeated until the growing peptide is completed.
- A termination codon is recognized by a release factor (RF), which induces peptidyltransferase to release the newly synthesized protein. The synthesizing complex dissociates; there is NO such thing as termination anticodon.
- Elongation of the polypeptide chain involves the addition of amino acids to the carboxyl end of the growing chain.
- During elongation, the ribosome moves from the 5’ -end to the 3’-end of the mRNA that is being translated.

14
Q
Drugs that disrupt translation
A
- Streptomycin - Inhibits initiation of prokaryotic translation
- Tetracycline, Puromycin, Chloramphenicol, Clindamycin, and Erythromycin all disrupt the prokaryotic elongation
- Diphtheria toxin disrupt the eukaryotic elongation
- Ricin - interferes with eukaryotic rRNA and essentially inactivates (exceptionally toxic)
15
Q
Polysome (polyribosome)
A
- More than one ribosome can proceed along the length of a mRNA at any time.
- Eukaryotic cell can have more than 1 million ribosomes in it
- RIbosomes move along the mRNA; the mRNA itself does NOT move