Transcription Flashcards
What is the main component of transcription factors that allows interaction with DNA?
The binding domain.
Binding Domain
Component of transcription factors that interacts with specific DNA sequences.
What do binding domains contain that form hydrogen bonds with DNA?
Structural binding motifs.
What are the DNA sequences that the binding domain interacts with sometimes called?
Elements.
What is the function of transcription factors?
To modify gene expression by interacting with DNA.
Transcription Factors
Proteins that interact with DNA to modify gene expression (transcription).
What kind of bonds do transcription factors form with bases in the DNA grooves?
Hydrogen bonds.
Are transcription factors exclusively activators or repressors?
Some are exclusively activators or repressors, but some are both.
<p>where do TF come from</p>
<p>-Some TF genes are constitutively expressed without the need for TFS while some TFs promote translation of other TFS</p>
<p>-TF are coded by other TF, some are coded by the GTC and are continuosly expressed.<br></br>-Many TF are inactive in cells and get activated by signals</p>
<p>-By being able to enter the nucleus from the cytoplasm.<br></br>Note Even if an inactive TF were placed in the nucleus it wouldn't work as its DNA binding sites are hidden away The same conformational change that allows it to pass through the nuclear pore also activates TF</p>
<p>Transcription Factor Activation points</p>
<p>-Protein Synthesis</p>
<p>-Ligand Binding</p>
<p>-Covalent modification</p>
<p>-Addition of a second subunit</p>
<p>-Dissociation from inhibitor</p>
<p>-Unmasking of bidning site</p>
<p>-Release from membrane</p>
What needs to happen for an inactive transcription factor to work in the nucleus?
Its DNA binding sites need to be unhidden.
What is a common covalent modification that can activate transcription factors?
Phosphorylation.
Transcription Factor Function Modification
Can occur when subunits bind to the same transcription factor.
What is required to activate expression of a single gene?
Multiple gene regulatory proteins (combinatorial control).
Combinatorial Control
Multiple gene regulatory proteins are required to activate expression of a single gene.
What must transcription factors do once in the nucleus?
Bind to the DNA and then either activate or repress transcription.
What type of bonds form between DNA bases and amino acid side chains?
Hydrogen bonds.
What can the binding of transcription factors cause in DNA?
<p>Changes in the structure of DNA that allows other components to interact with DNA and activate transcription</p>
<p>(change DNA shape to expose TATA).</p>
What is the role of TATA binding protein in transcription?
<p>Crucial for the assembly of the general transcription complex.</p>
<p>-its is a subunit of TFIID</p>
TATA Binding Protein (TBP)
<p>Part of TFIID that changes DNA shape to expose the TATA element for other components of TFIID to bind.</p>
<p></p>
Name three types of motifs that allow transcription factors to interact with DNA.
Helix-Turn-Helix, Zinc Finger, and Leucine Zipper.
Helix-Turn-Helix Motif
Common among homeodomain-containing transcription factors.
What is a homeodomain?
<p>A 60 amino acid sequence conserved across different transcription factors.</p>
<p>-TF structure may be different but the part that interacts With DNA remains the same</p>
What does the homeodomain encode?
A sequence that gives the helix-turn-helix structure.
A loop of twelve amino acids with conserved leucine and phenylalanine.
-Tetrahedrally co - ordinated zinc atom between the cysteine and histidine residues . - -Several basic amino acids project from the surface that interact with the DNA
Multiple zinc fingers interact with the DNA.
The number of zinc fingers within a transcription factor can vary- more fingers makes structure more complicated
Two transcription factors with leucine zippers that interact with DNA.
-Can be a homo or heterodimer
Acidic - high proportion of aa that give strong negative charge, Glutamine-rich, and Proline-rich.
How do activation domains change chromatin structure?
By recruiting and interacting with HAT and chromatin remodelling complexes.
-This causes structure of the DNA opens up to allow other transcription factors and the general transcription complex to bind to the DNA.
How do activation domains interact with the basal transcription complex
-Improving TFIID binding
• Enhancing the interaction between TFIIB and RNA pol II (helps position it on start site after TFIID binds)
• Interacting with TBP associated factors (TAFs) (aid in the recruitment of RNA Pol and other TFs)
-Bind to TBPs and interact with other TFs to recruit them to initiate the formation of the GTC
Links all transcription factors together to start transcription.
-It essentially facilitates communication between the transcription factors and the polymerase to ensure that transcription starts properly.
Interacting with activation domains and help recruit and stabilise components necessary for iniating transcription
Interact with activation domains to recruit other factors
Some TFs can't interact with TAFs by themselves.
-the co-factors enable binding which allows the TAFs to activate the transcrption factors
To travel and bind to TF as This usually occurs with large complex TF that are already bound to DNA
Repressor function
Can inhibit or prevent the function of activators.
-Repressors can directly repress gene expression.
-
To turn off gene expression but sometimes they can reduce experssion
Allows some TFs to be both activators and repressors.
R binds to binding site on gene instead of A- sometimes they have same structure while other times R has slighlty higher affinity
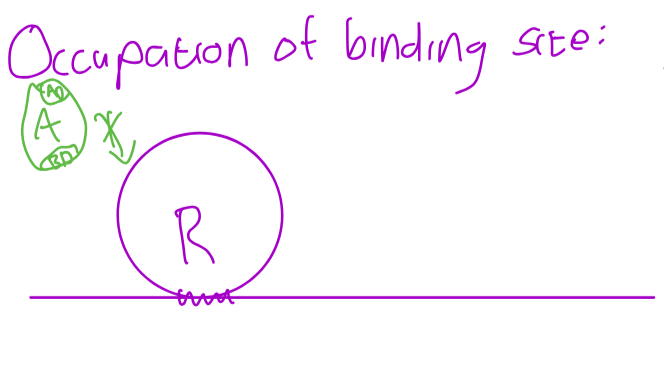
Repressor does not interact with DNA- Binds to binding domain of A
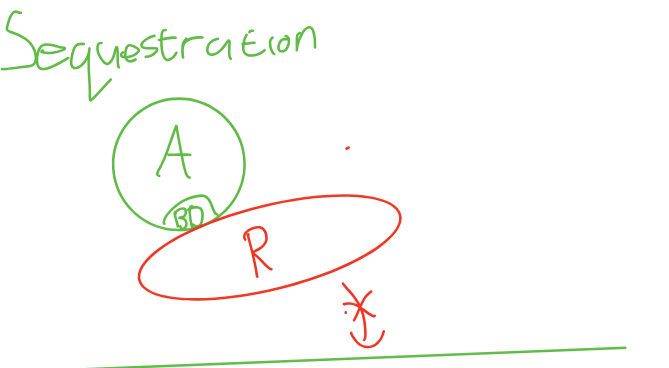
Activator binds to gene, but activation domain is blocked by R
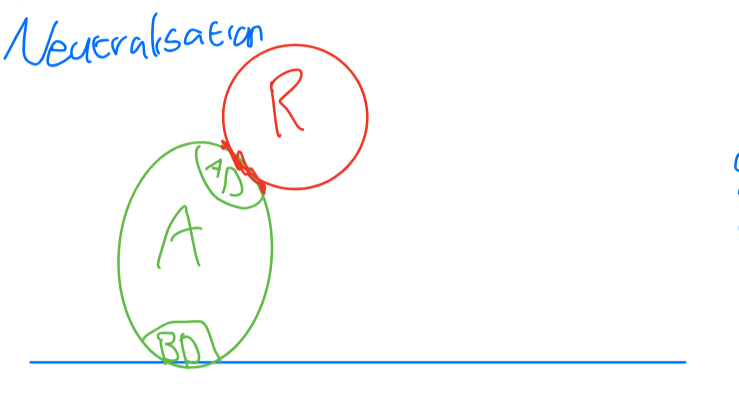
R adds ubiquitin groups to A (can occur in cytoplasm)
Proteasome recognizes ubiquitin groups and degrades activator.

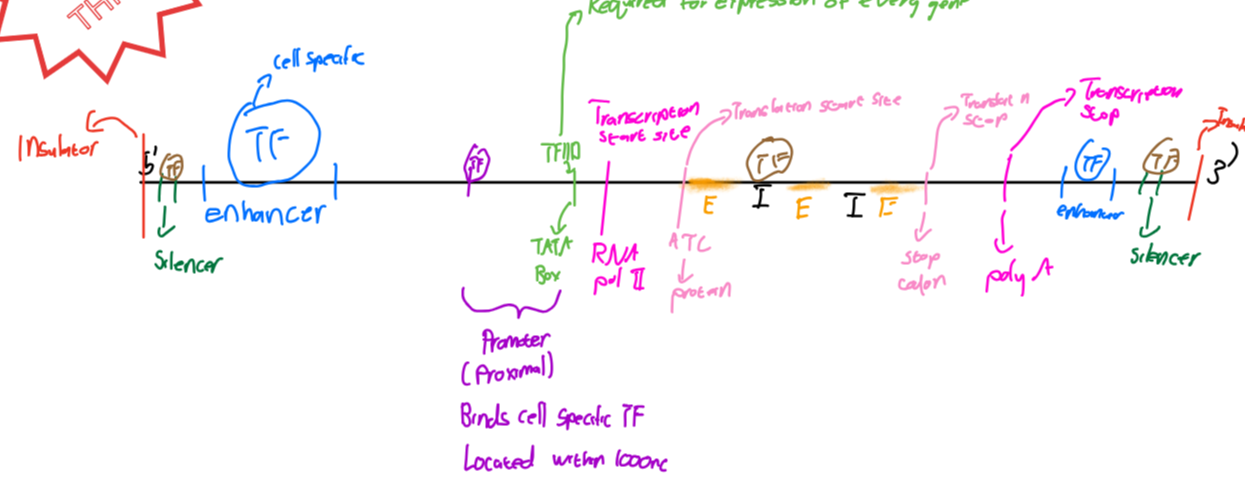
Basic structure of gene
Protein synthesis
-TF are made by other TF
-Some essential TF are made by GTC and are made every time
-Their regulation is very basic
Ligand binding
-Inactive TF where a ligand binds and causes a conformational change
-Many different types like copper or hormone (switches on Cu detoxifying gene)
Covalent modification
Phosphorylation is main one, also methylation, etc
-Works same way as ligand binding
Addition of a second subunit
-Inactive TF sometimes need an additional component like DNA binding or activation domain
-Usually occurs in TF already in nucleus
-Sometimes subunit is only synthesised due to signal
-Many subunits can bind to same TF which modifies its function
Dissociation from inhibitor
Many TF in cytoplasm are stuck to an inhibitor
-something needs to cause inhibitor to disassociate like a ligand or covalent modification
Unmasking of binding site
Unmasking of binding site
-Binding site needs to be exposed, same principle as inhibitor
Release from membrane
Anchored in cytoplasmic membrane and signal often extravellular causes it to be released. Signals themselves are too big to enter and so interact with receptors
TF and single and multiple gene expression
It can activate expression of many different genes, context specific.
-Multiple gene regulatory proteins activate expression of a single gene.
What structure of zn fingers sits in the major groove of DNA?
α-helix sits in the major groove while B sheet interacts with DNA backbone
Activation domains allow transcription factors to act as activators of transcription by:
-Interacting with components of basal transcription complex
-Interacting with mediator
-Interacting with SAGA
-Interacting with co-activators like CREB and CB
Direct Repression
Blocks binding of GTC
Blocks binding of chromatin modifiers
Degrades activators that are always present in case of emergency


