The bacterial genome Flashcards
How big are bacterial genomes?
Most prokaryontic genomes are 1 - 3 Mb large, however some are as large as 13 Mb (e.g. Sorangium cellulosum).
What is the main canonical frontier between cell world and virosphere?
Translation
What is special about the Tupanvirus?
It encodes largest translational apparatus within the known virosphere.
In this translation-associated gene set, only the ribosome is lacking.
What is special about the genome of pandoraviruses?
- Most complex viruses (genomes reach 2.5 Mb)
- Large fraction of the pan-genome codes for proteins without homologs in cells or other viruses
–> De novo gene creation could contribute to pandoravirus genomes
What is the traditional view of the bacterial genome?
- Bacterial genome is haploid
- the bacterial chromosome is a circular DNA molecule
- extrachromosomal DNA (pasmids) is circular and contain non-essential genes
- Bacterial chromosome is located in the “nucleoid”
–> Recent and ongoing genome projects challenge this view.
What is the nucleoid?
= region within the cytosol of bacteria that contains most of the DNA; can be easily visualized by staining methods
What is the definition of a bacterial chromosome?
de facto definition:
A chromosome is a DNA replicon that codes for house-keeping genes that are essential for the survival of the bacteria.
- Large DNA replicons are referred to as bacterial chromosomes
- Smaller ones are called extra-chromosomal elements, plasmids, or small chromosomes
- what if a small replicon is dispensable under lab conditions, but is crucial in the “real” world → should it be called plasmid or small chromosome?
- and how should one name replicons that are essential for life only under certain environmental conditions? Dispensable chromosomes?
–> all quite imprecise definitions
How does DNA Replication in bacteria work in general?
- Starts an origin of replication (ori)
- Is bidirectional
How many oris do the following organisms have:
E. coli, S. cerevisiae, H. sapiens
- E. coli: 1 ori
- S. cerevisiae: 300 oris (1 per 40 kb DNA)
- H. sapiens: 20’000 oris (1 per 150 kb DNA)
What problems does the replication of linear (bacterial) chromosomes pose?
- DNA replication of the 3‘ ends
- DNA polymerase doesn’t start de novo but can only extend an already existing strand (RNA primer)
BUT: RNA primer gets subsequently degraded → gap in the DNA strand
Eukarya solve this problem via telomerase
What proteins are collaborating at the replication fork?

- DNA helicase (brown mitten)
- Single-strand binding protein (4 black balls)
- RNA primase (green bell)
- DNA polymerase (orange donut)
What strategies can solve the problems associated with the replication of linear (bacterial) chromosomes?
Eukarya: solve this problem via telomerase
Bacteria:
- Borellia: ends of DNA double strand are covalently connected via hairpin loops
- Streptomyces: Special proteins are covalently connected to 5’ end → it is assumed that these proteins prime the terminal replication → Also known as Invertron Telomer
What is an Invertron Telomer and what does it do?
DNA polymerase interacts with the 5’-terminal protein (TP) and catalyzes the formation of a covalent bond between the TP and a dNTP. The dNTP bound to the TP has a free 3’-OH group which acts as the primer for chain elongation.

What is the general anatomy of a bacterial genome?
- Condensation to a bacterial chromosome is necessary to fit larger genome into smaller cell
- Done with supercoiling: happens when additional helical turns are introduced (positive supercoiling) or removed (negative supercoiling) within a circular DNA double strand
- DNA adopts the B-form helix
- Wide and accessible major groove, narrower minor groove
How does the structure model of the E. coli nucleoid look like?
- it is assumed that the E. coli genome is attached to a protein core structure from which about 12-80 supercoil loops emerge
- this genome organization is RNase-sensitive
- protein core consists of: DNA Gyrases & DNA Topoisomerases (needed for negative supercoiling & for relaxing positive supercoils; energy dependent process)
- most abundant proteins are the HU-proteins: 60-100 bp DNA are wrapped around one HU dimer (analogous to eukaryal histone proteins); ~60’000 HU molecules /cell → cover around 1/5 of the E. coli genome
How was the supercoil-loop model validated?
- radioactive radiation was used to introduce nicks into DNA and the effects on supercoiling were determined
- Supercoiling is monitored by TMP (Trimethylpsoralen) binding (binds better to relaxed DNA)
- if E. coli chromosome is not organized in supercoiled-loop-domains, then one nick should relax the entire chromosome
→ one would expect an “all or nothing” effect - However, a gradual, linear increase in TMP binding (and thus in DNA relaxation) was observed
–> supports supercoil-loop model
When can a bacteria be polyploid?
- In general the assumption that bacteria are haploid organisms is an oversimplification
- during exponential growth (especially in fast growing bacteria) > 4x sequence copies
close to the replication origins compared to the “ends” of the replicon (rRNA genes most often located close to the ori) - some species always carry multiple copies of their genome per cell (usually nearly identical copies)
- most of the times the advantage of polyploidy remains unclear, but most of the time it is a safeguard against mutations by gene conversion
Give three examples of bactreia that are polyploid.
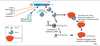
Deinococcus radiodurans:
- extreme resistant towards radiation and desiccation
- survives radioactive radiation of 17‘000 Gy –> 1’700x amount a human would die at
- has 5-8 copies of its chromosome/cell → oligoploid
- most likely needed to repair DNA strand breaks via homologous recombination
- Cells even survived low earht orbit (1 year outside ISS) and do not exhibit any morphological damage
- nano-sized particles over the surface of LEO-returned cells
- space-returned cells revealed pronounced outer membrane associated events with numerous vesicles
- metabolites, proteins and mRNAs were extracted from space-exposed cells
- proteome & transcriptome: multi-faceted response (e.g. UvrABC endonuclease excision repair upregulate; increased catalase & putrescine to cope with ROS)
Haloferax volcanii: halophilic archaeon
- Upon phosphate starvation, H. volcanii degrades its own gDNA to use the phosphate
- Ribosome concentration remains constant, thus rRNA is not used as P source
- Hypothesis: DNA might have evolved initially as storage polymer and only later gained its function as genetic material
- Can have over 30 genome copies/cell
Achromatium oxaliferum:
- Bacteria with multiple compartements, each containing own genomes
- Inside compartments, chromosomes might be independently replicated and use intracellular genen transfer to increase diversity
- Intermediate evolutionary state between uni- and multicellular life
- allow for the generation of “experimental” versions of functioning proteins or RNAs
What is gene conversion?
Asymmetrical homologous recombination resulting in one allele “overwriting” another
How does the E. coli genome look like?
- Genome is extremely compact
- possible advantage: faster replication time (e.g. during favorable environmental conditions).
- Only 11% of the E. coli genome consists of non-protein-coding DNA (H. sapiens > 90%)
- Both DNA strands are coding for genes
How is the complexity of an organism most likely determined?
Speculation:
the non-coding part of genomes is responsible for the increased complexity of e.g. mammals compared to bacteria
What is an operon?
- A characteristic hallmark of prokaryal genomes
- Quite frequently operons consist of genes that are involved in the same metabolic pathway
- e.g. Lactose operon or Tryptophan operon
- There are some bacteria that have operons that comprise of functinally unrelated genes
- e.g. Methanocossus jannaschii or Aquifex aeolicus
How does the gene order in bacterial genomes look like?
- Gene order in bacteria is very dynamic and not conserved
- Only weak similarity in gene order between bacterial phyla
-
But: Related operons have identical gene order (genes that work together, stay together)
- Reason: Horizontal gene transfer promotes clustering of genes that work together, since this increases chance to be fixed in the recipient genome
- Gene orientation often conserved
- Reason: To avoid head-on-collisions of the replication and transcription machineries
What is special about bacterial RNA transciption?
- Bacterial transcripts are typically uncapped
- Also some bacterial RNAs are capped:
- Bacterial coenzyme-cap added during initiation
- Bacterial cap provides RNA stability (other roles?)