Mark Isalan Flashcards
(165 cards)
General organisation of DNA in Eukaryotes?
Eukaryotic DNA is Heavily organized
DNA highly condensed to form chromosome –> organization driven by histone complexes which are positively charged so that they can interact with the negatively charged DNA
In the highly supercoiled form the DNA is not accessible to transcription factors and other enzymes

General organisation of DNA in Bacteria?
Even though the Bacterial genome is less organised than eukaryotic DNA. bacterial genome is not organised randomly
DNA is still has heavily organised supercoiled circular nucleoids –> Refers to DNA is associated with several proteins such as Histone-like nucleoid structuring (H-NS) protein (positively charged) as well as other proteins such as transcription factors.
General Overview of DNA synthesis?
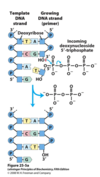
DNA synthesis - Occurs in 5’ to 3’ direction
Remember that DNA has polarity and is anti-parallel
5’ end has a free phosphate
3’ -OH has a free -OH
Synthesis of more DNA requires DNA polymerase (works in a 5’ to 3’ direction) which uses ATP, Mg++ and nucleotide triphosphates
During synthesis of the 3’ -OH performs a nucleophilic attack on the incoming deoxynucleotide triphosphate in order to add the base pair to the growing chain
Product - Add Nucleotide + Disphosphate released
Is DNA a stable molecule?
No clear answer
- In one sense yes, as it can last for 100’s of years in permafrost –> providing readable DNA
- In another sense no, as DNA is always very active –> replication, transcription, unwinding –> resulting in many sources of damage and error.
What are mutagens?
Mutagen –> Refers to a physical (i.e. U.V.) or chemical (i.e. Free radical species) agent that changes the genetic material
How frequently is DNA damaged in one cell per day?
DNA is damaged approximately 10 000 times per cell per day –> all of which act as a source of potential diseases/disorders
Hence, this explains why the cell devotes a lot of energy to prevent DNA damage
Two main impacts of DNA damage?
- Block replication and/or transcription
- Cause alterations in the genetic code (mutation) –> impact the organism itself or offspring if germline cells are mutated.
What are the two main types of DNA damage?
- Chemical alteration to DNA –> which may be exogenous** (source of damage located outside the cell - environmental mutants such as UV radiation) or **endogenous (internal source of damage - internally generated damaging agents such as hydroxyl radicals – most common)
- Spontaneous damage to DNA –> DNA reacts with itself Includes deamination (losing an amine group) and depurination (losing a purine base) –> these changes tend to occur regardless of what you do, meaning they are inevitable.
Outline how UV may lead to the formation of pyrimidine dimers (exogenous - chemical change).
Example of exogenous agent causing DNA damage
- UV light induces formation of pyrimidine dimers (T-T (most common) C-T and C-C) –> 2 adjacent pyrimidines are joined by a cyclobutane(4C) ring structure (Double C=C converted to single and remaining electron forms C-C bond with adjacent pyrimidine)
- Consequence - The bases no longer function as normal Watson and Crick base pairing –> no longer serve its function as a base - no information
This explains why Solar UV irradiation is the cause of most human skin cancer.
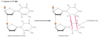
Outline how DMS/EMS (Exogenous) causes DNA damage.
DMS/EMS both are alkylating carcinogens
Alkylation is the addition of methyl or ethyl groups to various positions on the DNA bases
For example,
Alkylation of the O6 position of guanine results in formation of O6-methylguanine –> this changes the W-C base pairing potential. Normal G forms three H-bonds but this no longer occurs when carbon 6 has become methylated.
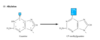
Is normal cellular DNA methylation a source of DNA damage?
No! Different to the alkylation due to DMS/EMS
Normal methylation from the cell is normally not carcinogenic
Takes place on CPG islands and effects the major groove whereas carcinogens disturb the Watson and Crick base pairing.
Outline how benzo-(a)pyrene (exogenous agent) can damage DNA.
Many carcinogens (e.g.benzo-(a)pyrene) react with DNA bases, resulting in the addition of large bulky chemical groups to the DNA molecule
Result? –> messing up the normal W-C base pairing –> Base is misread by DNA polymerase (as thymine)
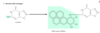
Are many carcinogens activated endogenously?
Yes!
Many carcinogens are activated endogenously, become free radicals, by reactions with cytochrome P450 enzymes
P450-important in clearance of compounds – ‘molecular dustbin’ –> may result in the formation of reactive intermediates.
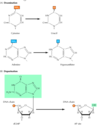
What are the two main types of Spontaneous Damage that occur in DNA?
A) Deamination of adenine, cytosine and guanine –> E.g. Amine to carbonyl –> change from hydrogen bonding potential (donor –> acceptor)
Deamination examples - Cytosine to Uracil and Adenine to Hypoxanthine –> changes the W-C base pairing potential.
(B) Depurination –> removal of purine group –> resulting from cleavage of the bond between the purine bases and deoxyribose, leaving an apurinic (AP) site in DNA.
Depurination - losing an entire base –> leaves sugar behind with empty –OH group –>
Note - same thing can happen with pyramidines (depyrimidination)
What does changed W-C base pairing potential mean?
Basically, DNA damage leads to altered base pairing - chemical change to base results in difference base preference (Non-WC base pairing) - when it gets replicated the wrong complement base gets included.
Example
Adenine deamination changes the C-NH2 on carbon 6 to C=O, which changes the molecule from adenine to Hypoxanthine.
Hypoxanthine behaves more like guanine therefore it preferentially binds to cytosine.
Thus, in the short term Hypoxanthine does not bond well with Thymine creating a bulge in the DNA.
In the long term, during DNA replication hypoxanthine will bind to cytosine –> resulting in a permanent mutation which may result in deleterious effects

What are common functional groups found in carcinogens?
Functional groups in carcinogens?
It can be a variety but all of them are reactive groups –> One common example would be aldehydes
Sometimes carcinogens aren’t even covalently reacting - e.g. intercalating agents sit in between the bases –> disrupting the DNA synthesis resulting in the wrong base being introduced –> e.g. ethidium bromide
What are the two general types of DNA repair mechanisms?
- DIRECT REVERSAL of chemical reaction responsible for DNA damage (in-situ) –> more common in bacteria
- EXCISION REPAIR –> Removal of damaged bases, replacement with newly synthesised DNA more common than direct repair in humans
Why? Because humans often lack some of the enzymes required for direct reversal
Outline the direct reversal of U.V. DNA damage in E. Coli?
- DNA damage via U.V. radiation
- Pyrimidine dimer created (cyclobutene rings)
- Photoreactivating enzyme uses photoreactivation (process driven by light as energy source) to break open cyclobutene ring to restore normal bases
How does the photoreactivating enzyme do this?
- Enzyme has a chromophore that can absorb light 300nm-500nm
- Transfers the energy to a non-covalently bound FADH-, which then transfer an excited electron to the pyrimidine dimer –> in turn splitting the ring.
- The now pyrimidine anion transfer its electron to FADH. (Radical) making the DNA good as new.
Note - Occurs in E. coli, yeasts, some plant and animal cells but NOT in humans
Outline how dealkylating enzymes can be used against the direct reversal of DNA damage.
Dealkylating enzymes –> removes alkylating group - enzyme has methyl acceptor group (–SH group)
- Enzyme comes in
- Finds bulge in DNA
- pulls of methyl group using –SH
- Base returns to its original form
- Enzyme needs to be regenerate using other proteins (reform –SH)
Specific Example
Alkylation - O6-methylguanine –> How can we fix?
- Can be repaired by enzyme: O6-methylguanine methyltransferase which is widespread in prokaryotes and eukaryotes
- Transfers the methyl group to its own cytosine residues -> reaction deactivates the protein which therefore cannot be strictly classified as an enzyme.
Types of Excision Repair?
- Base-excision repair –> Smallest type of excision - Base is removed leaving deoxyribose backbone intact so that another series of enzymes can add the correct base
2. Nucleotide-excision repair –> Nucleotide is removed leading to a gap in one strand (an oligonucleotide is usually removed) which is filled in by DNA polymerase
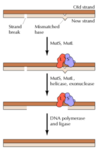
- Mismatch Repair –> Mechanisms for post DNA replication mismatches.
Outline how base excision repair is used to fix deamination of cytosine to uracil.
Base excision repair example - Convert Uracil back to Cytosine
- Uracil formed by deamination of cytosine, leads to a G:U mismatch –> results in DNA bulge
- Bulge is recognized by DNA glycosylase resulting in the bond between uracil and deoxyribose being cleaved by uracil DNA glycosylase –> leaves a sugar with no base attached in the DNA (an AP site)
- This site is recognized by AP endonuclease (cuts into intact DNA), which cleaves/nicks the DNA chain
- The remaining deoxyribose is removed by deoxyribose-phosphodiesterase –> leaves perfect gap for new nucleotide
- The resulting gap is filled by DNA polymerase and sealed by ligase - leads to incorporation of C opposite G.

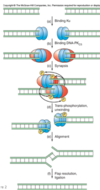
Outline how nucleotide excision repair of thymine dimer (UV damage)
Nucleotide excision repair of TT dimer - Main mechanism in Humans
Note - This also occurs in many organisms or alternatively they use photo repair enzymes.
- Recognition of thymine dimers occurs by assembly of RPA, XPA and XPC-TFIIH at sites of damage - specificity is achieved mainly by the kinetic proofreading activity of TFIIH
- Helicase action by the XPD subunit of TFIIH generates a bubble around the dimer - creating the requisite DNA substrates for the structure-specific endonucleases XPF and XPG
- Cleave on both sides of the thymine dimer by 3’ and 5’ endonucleases creating nicks - XPF and XPG
- Oligonucleotide 24–32-nt in length (also known as the “canonical 30-mer”) dissociates from the duplex.
- The resulting gap is then filled by DNA polymerase and sealed by ligase (DNA pol I in E. coli/DNA pol β in human)
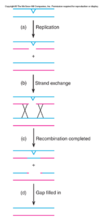
How does Nucleotide-excision repair differ between E. Coli and Eukaryotes?
-
E. Coli - Catalysed by 3 gene products – uvrA, B, C
a) UvrA recognises damaged DNA (Helix distortion),
b) UvrB and UvrC (endonucleases) cleave at 3’ and 5’ sides, excise 12-13 bases oligonucleotide
- UvrA, UvrB and UvrC form complex
c) uvrD (Helicase II) binds to the oligonucleotide segment in order to displace it –> allowing for DNA pol and DNA ligase binding.
- Mutations of these genes leads to high sensitivity to UV - Eukaryotes - Catalysed by RAD gene products in yeast (7 different repair genes involved – highly conserved) –> analogous to humans
Mutations lead to xeroderma pigmentosum rare genetic disorder, affects 1:250,000 people, extreme sensitivity to UV light leading to skin cancers - all of which is due to the deficient ability to repair DNA by nucleotide-excision.
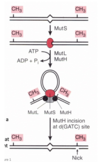
In Mismatch repair in E. coli, how does the E. Coli strand distinguish between parental and new synthesized DNA strand?
Main idea - Mismatch repair system detects and excises mismatched bases in newly replicated DNA
Important characteristic is that this system must distinguish parental strand from newly synthesised daughter strand because you want to change the base in the new strand (not change genetic code on the parental strand)
Different species use different methods to accomplish this…
In E. coli DNA is methylated by Dam methylase (Adds methyl group to Adenine in GATC) –> following replication the newly synthesised daughter strand will not be methylated resulting in hemi-methylated DNA –> so the cell can recognize any bulge on any unmethylated new DNA.