Lecture 2: DNA Replication, Recombination and Repair Flashcards
(32 cards)
_________ are necessary for long term survival of a species.
What is a mutation? Silent mutation?
Low mutation rates are necessary for long term survival of a species.
Mutation is a permanent change in DNA. Can be deleterious or silent (which is when it changes a codon but not the AA, or the AA changes but doesn’t affect protein function.
What is the mutation rate for DNA in humans?
1 in every 109 nucleotides each time DNA is replicated (so about 3 nucleotides each time DNA is replicated)
Which enzyme catalyzes DNA synthesis?
How does the conformational change of this enzyme affect DNA synthesis?
DNA polymerase catalyzes DNA synthesis.
Think of DNA polymerase as a hand. The deoxynucleoside triphosphate will come in, the base pairs will line up and hydrogen bond, the correct base pair creates a large favorable change in free energy, the “hand closes”, once pyrophosphate comes off, the hand opens back up again and allows the strand to translocate

The DNA replication fork is ______
What does this allow to happen?
DNA replication fork is asymmetrical
This allows DNA synthesis to occur in the 5’ to 3’ end (it always occurs in the 5’ to 3’ direction)
Able to synthesize both daughter strand simultaneously (both leading and lagging strand)

What are Okazaki fragments?
They are fragments of DNA made by a primer for the lagging strand. They get joined together by DNA ligase. These are important because they facility 5’ to 3’ synthesis of DNA and FIDELITY can be maintained
What are the three proofreading mechanisms for DNA synthesis that allow high fidelity of DNA replication?
- 5’ to 3’ polymerization
- The correct nucleotide has higher affinity for the moving polymerase
- After base pair binding, polymerase must undergo conformational change which happens a lot easier if its the correct nucleotide
- Exonucleolytic proofreading in 3’ to 5’ direction polymerase can be self correcting
- Strand directed mismatch repair

Describe how DNA is synthesized on the lagging strand.
DNA primase creates RNA primers.
DNA polymerase then comes in and finishes the DNA fragments.
The RNA primers get removed, but then there are nicks in the DNA strand.
Those nicks are fixed by DNA ligase.

What do helicase and single stranded DNA binding proteins required for?
Explain helicase and what it does. What kind of DNA does it move down?
Explain single stranded DNA binding proteins. What do they bind to?
Helicase and single stranded DNA binding proteins are required to open up the double helix ahead of the replication fork.
Helicase: moves down single stranded DNA and opens up double stranded DNA… it involves ATP hydrolysis and moves in a 5’ to 3’ direction
ssBPs: these stabilize the single stranded unwound DNA through cooperative protein binding. They bind to the sugar-phosphate backbone so that they don’t cover up the base pairs and get in DNA poly’s way.

What is the purpose of the “sliding clamp”?
The sliding clamp holds the DNA Polymerase on the DNA (otherwise it would fall off).
Important: the sliding clamp falls off once DNA polymerase gets to double stranded DNA
Give a summary of the DNA replication machinery that takes place at the replication fork. You can use the figure below to guide you.
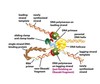
At the front of the replication fork, DNA helicase opens the DNA helix. • Two DNA polymerase molecules work at the fork, one on the leading strand and one on the lagging strand. • Whereas the DNA polymerase molecule on the leading strand can operate in a continuous fashion, the DNA polymerase molecule on the lagging strand must restart at short intervals, using a short RNA primer made by a DNA primase molecule. • A folding back of the lagging strand makes the close association of the subunits possible. • This arrangement also facilitates the loading of the polymerase clamp each time that an Okazaki fragment is synthesized: the clamp loader and the lagging-strand DNA polymerase molecule are kept in place as a part of the protein machine even when they detach from their DNA template. • On the lagging strand, the DNA replication machine leaves behind a series of unsealed Okazaki fragments, which still contain the RNA that primed their synthesis at their 5’ ends. This RNA is removed and the resulting gap is filled in by DNA repair enzymes that operate behind the replication fork
How does strand directed mismatch repair work?
The strand directed mismatch repair system detects the distortion in the DNA helix caused by the misfit of noncomplementary base pairs.
There is a protein complex of MutS and MutL. Mut S binds to the mismatch. MutL scans for a nick. Then the complex of MutS and MutL create a loop. That loop gets clipped out. DNA Polymerase then comes in and completes the synthesis.

Which enzyme prevents DNA tangling during replication?
DNA topoisomerase
What is the difference between Bacterial DNA vs Eukaryotic DNA when it comes to origins of replication?
During what cell cycle phase is DNA replicated in Eukaryotes?
Bacterial DNA only contains one origin of replication, whereas eukaryotic DNA contains several origins of replication
DNA is replicated during S phase
What happens to nucleosomes behind the replication fork?
What are CAF 1 and NAP 1 and what do they do?
New nucleosomes are assembled behind the replication fork.
CAF1 and NAP1 are histone chaperones.
CAF1 loads newly synthesized H3-H4 tetramers
NAP1 loads H2A-H2B dimers

What happens with modified histones on DNA after replication?
Well, after replication, only half of the daughter nucleosomes will have modified histones. So, to fix that, reader writer complexes will come in and re-establish the parental pattern (EPIGENETICS)

Telomerase gives you a repeating structure at the end of chromosomes that then loop back together to form a “________”
What does that specialized structure do/why is it important?
“T Loop”
T loops give normal ends of chromosomes a unique structure. T loops protect the ends from degradative enzymes. T loops also help distinguish normal chromosomes from ends of broken DNA molecules.
Telomeres also have a role in cell _____ and the prevention of _____.
Telomeres have a role in cell senescence and the prevention of cancer.
__________ & ________ are the most frequent spontaneous chemical reactions known to cause DNA damage.
Depurination and deamination
Depurination: legit removing a guanine or adenine
Deamination: example is changing cytosine to uracil

What are the effects of depurination and deamination?

UV radiation causes what kind of DNA damage?
UV radiation creates dimerization of two neighboring pyramidines. (C-C, T-T, C-T)
What are the two major DNA Repair Pathways?
Base Excision Repair, and Nucleotide Excision Repair
Explain the mechanism for base excision repair
So we have a messed up nucleotide (example is deaminated C).
- The first step is that a glycosylase will remove the base that screwed up
- Then, AP Endonuclease and Phosphodiesterase will remove the sugar phosphate
(now you have DNA with a single nucleotide gap)
- DNA polymerase will add the new nucleotide, and DNA Ligase will seal the nick
Explain the mechanism for Nucleoside Excision Repair
So we have something that is screwed up (example given in class is pyramidine dimer)
- Excision Nuclease marks recognition sequence and cuts it off (kinda far from the issue, cuts a whole chunk)
- Helicase will unwind that segment and it will go away
(now we have DNA with a long gap, like 12 nucleotide gap)
- DNA Poly will come in and synthesize the complement from 5’ to 3’ to fill the gap, and DNA Ligase will seal the nick
What ensures that the cell’s most important DNA is efficiently repaired?
Coupling DNA repair to transcription


