Key notes Flashcards
(43 cards)
1
Q
- There exist many computational methods that are available depending on the length/time scale we are interested in.
- Classes in order of increasing … scale and decreasing … scale they investigate
- Quantum/… … : includes atoms, electrons in an explicit solvent using … … (Schrodinger). Accurate but computationally unfeasible for >… atoms.
- … -…: all/most atoms included in an explicit solvent but instead uses…as a method of reference, which describe atoms via….
- Coarse-grained: group 4-5 heavy atoms into beads, in an explicit or … solvent using MD. Far less interactions to compute but at the cost of accuracy.
- … -…-… : interaction sites grouped, comprising of many atoms, generally proteins/peptides, using an implicit solvent with … dynamics.
- … : Materials represented as a continuous mass in an implicit solvent, using … … (instead of QM/EOM).
A
- There exist many computational methods that are available depending on the length/time scale we are interested in.
- Classes in order of increasing length scale and decreasing time scale they investigate
- Quantum/Ab initio: includes atoms, electrons in an explicit solvent using quantum mechanics (Schrodinger). Accurate but computationally unfeasible for >3 atoms.
- All-atom: all/most atoms included in an explicit solvent but instead uses MD as a method of reference, which describe atoms via EOMs.
- Coarse-grained: group 4-5 heavy atoms into beads, in an explicit or implicit solvent using MD. Far less interactions to compute but at the cost of accuracy.
- Supra-coarse-grained: interaction sites grouped, comprising of many atoms, generally proteins/peptides, using an implicit solvent with stochastic dynamics.
- Continuum: Materials represented as a continuous mass in an implicit solvent, using continuous dynamics (instead of QM/EOM).
2
Q
- Define rare events in the context of molecular simulations.
A
- Processes that involve time scales much longer than what we can computationally afford.
- Definition relative to our method of choice.
3
Q
What are enhanced sampling techniques
A
- Computational methods that allow one to overcome the timescale problem by sampling the phase space of our system to a greater extent. We can use MD or MC to do this.
4
Q
- (IMP) How is system free energy related to the timescale problem.
A
- Our phase space if a collection of all configuration’s position and momenta.
- A rare event is likely to be separated from our starting configuration by a high free energy barrier that cannot be overcome using MD/MC alone in a feasible timescale. Instead we are likely to spend all/most of our simulation time trapped in that local minima.
5
Q
- (IMP) Briefly describe the two main methods of enhanced sampling.
A
- Free energy-based methods: characterize PES of system
- E.g. umbrella sampling, replica exchange MD, metadynamics
- Choice of reaction coordinate/dof important and will affect free E of system.
- No direct info about kinetics of the process
- Path sampling-based methods: explore all possible pathways
- E.g. transition path/interface sampling, forward flux sampling
- More suited for processes with many possible pathways between
- No direct info about the thermodynamics of the system.
6
Q
- What is an order parameter?
A
- An order parameter or collective variable, allows the different configurations/states of a system to be distinguished
- Allows system to be driven from A to B (or v.v) through application of enhanced sampling method of choice.
7
Q
- What are the requirements of an OP? what is the difficulty in this requirement?
A
- Must be differentiable with respect to atomic position
- This is simple in the cases illustrated but can be very challenging for complex systems that is simple enough to compute but accurately represents dynamics.
- Can also be difficult when no prior knowledge of configurational space is known.
8
Q
- What is affected by our OP choice in free energy-based methods.
A
- Our choice of OP determines the resulting free energy surface (FES)
- This multidimensional hyper surface is re-mapped on to a simple coarse-grained FES (1D/2D)
9
Q
- What is problem with this new FES?
A
- Can often be an oversimplification as only representative of the chosen order parameter.
- The true FES, which represents all configurational space does not equal the FES we are sampling, constrained to our OP of choice
10
Q
- (IMP) How does statistical mechanics relate to our FES assumption?
A
- Stat mech confirms true FES ≠ our FES according to this OP, as uses configurational partition function, Z which does not contain info about kinetics/ dynamics of the system and depends only on particles position in the system only.
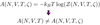
11
Q
(PPQ) How can we use commitor analysis to probe the accuracy of our OP?
A
- Select a value, OP* of OP corresponding to a putative transition state (maximum)
- Sample an ensemble of configurations characterized by OP*
- Run several MD simulations for each, varying velocity (making them statistically independent)
- Find probability of OP* configuration making to it to B
- Plot histogram of these probabilities.
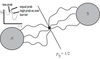
12
Q
- (PPQ) Which of these histograms indicates a more accurate OP?

A
- (a) indicates no matter where you start you always end up with same probability of ending up in A or B, NO indication this is a TS
- (b) indicates probability of ending up in B increasing as you move further along coordinate, 0.5 at middle. Representative of the putative TS
- Therefore, b is better.
13
Q
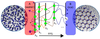
(IMP)
- … … … is a path based enhanced sampling method where instead of computing an overall (v.low) … , path is divided into a series of …
- Each … has an increasing value of our …, … each corresponding to a possible … with said value of …
- As λ increases likelihood of going from … to … , rather than back to A, …
A
(IMP)
- Forward flux sampling is a path based enhanced sampling method where instead of computing an overall (v.low) probability, path is divided into a series of interfaces
- Each interface has an increasing value of our OP, λ, each corresponding to a possible configuration with said value of λ
- As λ increases likelihood of going from A to B, rather than back to A, increases
14
Q
(IMP) How are trajectories generating in FFS?
A
- Begin by looking at fluctuations of a long unbiased MD run
- At each interface λi large # of trial molecular dynamics runs done
- The few that reach the next interface λi+1 are used as a starting point to reach the following interface.

15
Q
(IMP)
- ML methods are good at … data, but poor are … predictions.
- In other words, ML can give accurate prediction … … … … (filling the gaps), but poor at predicting data … of it.
- The … /… of data accessible and how we … it is much more important than the algorithm we feed it in to.
A
(IMP)
- ML methods are good at interpolating data, but poor are extrapolating predictions.
- In other words, ML can give accurate prediction within a data set (filling the gaps), but poor at predicting data outside of it.
- The amount/quality of data accessible and how we describe it is much more important than the algorithm we feed it in to.
16
Q
Why is there a need for ML-based interatomic potentials in simulations?
A
- Classical forcefields lack the detail to accurately reproduce complex systems, as functional forms can be very limiting.
- The timescales these systems exist in are also far too large for quantum calculations.
17
Q
- What are the pros and cons of ML-based interatomic potentials?
A
- Pros: fast, long large scale simulations with quasi quantum chemistry accuracy (if data set is good
- Cons: Not easy to craft dataset, can take years to improve iteratively.
18
Q
What is the process in which ML is used in drug discovery
A
- Dataset of small molecules attained
- Descriptors used to encode dataset of structures that can be processed by an algorithm.
- ML algorithm used to optimise dataset
- Desired property (e.g. solubility) predicted
19
Q
- What is a descriptor?
A
- A mathematical object (vector) that contains information about the system, encoded to be readily fed into a ML algorithm. Often, must satisfy specific properties like permutation invariance (same descriptor when exchanging identical atoms)
- Is the most crucial step in ML drug design
20
Q
- Give an example of a simple and a complex descriptor
A
- Simple: molecular weight
- Complex: Largest eigenvalue of adjacency matrix, where 1 is connected and 0 is not, giving a unique # corresponding to a unique matrix.
21
Q
- What is the main problem with ML?
A
- Understanding the interactions underpinning the PES of a system (ML potentials) or structure function relation between drug and potency (ML drug discovery) is very difficult as ML is a black box with many hidden layers between input and output.
22
Q
(IMP) Label this neural network
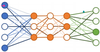
A

23
Q
- What are weights in a neural network and how do they aid the formation of further layers?
A
- Weights are numbers assigned to input nodes to form the first hidden layer.
- The value of y of the third node in the first hidden layer is a linear combination of descriptors and all connected weights
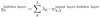
24
Q
- How is data fed in to a neural network?
A
- Dataset generated (e.g. N molecules and their solubility values)
- Descriptor values assigned to molecules (e.g. largest eigenvalue λimax of the adjacency matrix of the ith molecule)
- Values represent input neurons/nodes forming the input layer
25
(PPQ) What is the role of the activation function in artificial NN’s, give an example of one to support your choice?
* Activation function are non-linear functions (e.g. the sigmoidal function) that transform the linear combinations into non-linear objects from one hidden layer to the next.
* These linear combinations (e.g. relation between descriptors and solubility) are smoothed out, giving non-linear capabilities
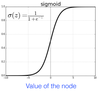
26
Give the general equation describing the value a node in the second hidden layer in a NN
* Activation function only present in the hidden layers (=1 for input)
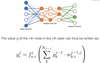
27
(IMP) How can we solve the issue of poor output values due to initial input weights?
* Use Backpropagation, where a cost function, fcost is calculated.
* This is the sum of output layer errors and target values (e.g. experimental solubility), written in terms of weights.
* Its derivatives are used to improve initial guess of weights to assign to minimise fcost on next iteration.

28
(IMP) What is the purpose of the hidden layers?
* Connect the results of our input nodes and weights and combines them further additional layers to fully optimise all input values.
* More hidden layers, more nodes, more flexible functional form is, up to the point where overfitting begins
29
* An **...** represents each set of output values generated.
* At the end of each **...** we compute the **...** **...** and use its derivatives to optimise the **...**
* We stop this when our model is good enough that the **...** value of our molecule of choice generates an accurate enough output value.
* An **epoch** represents each set of output values generated.
* At the end of each **epoch** we compute the **cost function** and use its derivatives to optimise the **weights.**
* We stop this when our model is good enough that the **descriptor** value of our molecule of choice generates an accurate enough output value.
30
What are Gaussian processes?
Mathematical objects which can be used to fit data through regressions via the generalization to infinite dimensions of a normal Gaussian distribution.
31
(IMP) What are Kernel functions (K)?
* The **covariance matrix** of our GP defines the **shape of ensemble of Gaussians** in space.
* A **functional form** for it must be written in terms of **hyper-parameters that can be optimised.**
* This mathematical expression is required as the covariance is an arbitrary set of numbers in a matrix
* For each element i,j of the covariance matrix can write an expression called a kernel, which is a function of the xi, xj descriptor point yi, yj in our dataset.
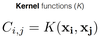
32
(IMP) Choosing the Kernel functional form can be very challenging. Give an example of a common choice
* The radial basis function (RBF) kernel
* A measure of the similarity between the two descriptors (i.e. between two molecules), as it is a function of the distance between them
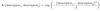
33
(IMP)
* The hyper-parameter L is the quantity **...** according to the **...** **...** **...** to obtain our ML model using **...** **.**
* It quantifies the **...** **...** by which two descriptors are close or not, giving the **...** of the resulting GP.
* Like weights in NN’s
(IMP)
* The hyper-parameter L is the quantity **optimised** according to the **log marginal likelihood** to obtain our ML model using **GP’s.**
* It quantifies the **length scale** by which two descriptors are close or not, giving the **smoothness** of the resulting GP.
* Like weights in NN’s
34
* (IMP) What is overfitting in ML
* After **many regressions** our **error** with respect to the cost function of our log marginal likelihood is very **small** giving numerically sounds results relative to our input data.
* However, will reach a point where so much fitting is done that a resulting graph like L=0.3 forms which gives very poor predictions in practice.
35
(IMP) How can we solve the issue of overfitting?
* Split our data into training and test sets.
* The training set (~80%) is used to build a model and the test set (~20%) is used to evaluate its predictive capabilities.
36
(IMP) What would overfitting indicate about training/test data
* If error in training set is very small and the error in its predictive capabilities is high, the data is overfit.
* The opposite would indicate underfit data.
37
(IMP) Why is it easier to spot overfitting in NNs than GPs?
* NN split in to epochs, so when divergence in error between error in test and training data occurs, can move back to epoch before this.
* Overfitting in GPs more difficult to detect/fix.
38
* Good numerical **...** of ML model with respect to the training set does NOT ensure the **...** of its **...** capabilities (IMP -sketch)
* Even if going through all points **(****...**≈ 0),**...** in between these points may be large,
* Good numerical **accuracy** of ML model with respect to the training set does NOT ensure the **accurac**y of its **predictive** capabilities (IMP -sketch)
* Even if going through all points **(fcost** ≈ 0), **error** in between these points may be large,

39
* What is the process of breaking down a molecular structure in to descriptors we can feed in to our ML model?
* Convert 3D structure to 2D
* Decompose 2D structure in a way that can be made into an adjacency matrix
* Diagonalize this matric to get eigenvalues that can be used as a principle eigenvalue in descriptors
40
* What are cliques and why are they useful?
* Cliques are the subunits that comprise all the molecules in our training set, allowing us to make a CG rep of each molecule of these clique components.
41
* One hot encoding is a process by which categorical variables are converted into a form that could be provided to ML algorithms to do a better job in prediction
* Each jth clique is represented by a **...** containing **...** elements i.e. in 100 cliques each clique is represented by a **...** with **...** elements
* These Nclq **...** are all equal to **...** except one; the one element corresponding to the j-th clique.
* A molecule is represented by the **...** of all its cliques.
* One hot encoding is a process by which categorical variables are converted into a form that could be provided to ML algorithms to do a better job in prediction
* Each jth clique is represented by a **vector** containing **Nclq** elements i.e. in 100 cliques each clique is represented by a **vector** with **100** elements
* These Nclq **elements** are all equal to **0** except one; the one element corresponding to the j-th clique.
* A molecule is represented by the **sum** of all its cliques.
42
* What is a pro and con of using this coarse-grained representation
* Pro: Highlights the importance of different functional groups, reducing noise
* Con: sacrificing some detail, which scales poorly with the amount of data being used.
43
(IMP)
* Once **...** cannot be improved anymore, choice of **...** can be tuned to best suit **...**
* ARD **...** uses a **...** of different **...** **,** one for each **...** in ensemble. This is useful as different **...** can have different **...** **...**.
* Gives us an idea of which **...** matter most.
* Once **descriptor** cannot be improved anymore, choice of **kernel** can be tuned to best suit **descriptors**
* ARD **kernel** uses a **combination** of different **kernels,** one for each **descriptor** in ensemble. This is useful as different **descriptors** can have different **length** **scales**.
* Gives us an idea of which **descriptors** matter most.


