Histology / Cell Biology Flashcards
What is angiogenesis?
The growth of new blood vessels from preexsiting vessels
Tumor cells ___ ___invade normal tissue and capture existing blood vessels, rather tumor cells ________ normal endothelial cells to form new blood vessels.
Tumors DO NOT invade normal tissue and capture existing blood vessels, rather tumors recruit normal endothelial cells to form new blood vessels
Describe the general process of angiogenesis
Cells typically monitor Oxygen (O2) tension. Under hypoxic conditions cells release angiogenic factors such as VEGF (vascular endothelial growth factors). VEFG binds to tyrosine kinase receptors on the surface of endothelial cells stimulating their proliferation. Endothelial cells migrate towards areas of higher VEGF concentration. In this way, VEGF can act as a chemoattract.
What is the angiogenic “switch”?
- Normal conditions prevent cells from triggering angiogensis
- As tumors progress they gain the capacity to promote angiogenesis
- Thus, overcoming the normal inhibition of angiogenesis is a step or switch, in tumor progression
What are the differences between normal capillaries and tumor capillaries?
Tumor capillaries are:
- Larger - 3 times the diameter
- Disorganized layout
- Loosely associated pericytes
- Gaps of several microns between cells
- Permeable - Walls up to 10 times more permeable
- High Pressure - Higher interstitial fluid pressure
- Poor Drainage - Poor lymphatic drainage
Name some angiogenic growth factors
- VEGF (Vascular Endothelial Growth Factor)
- TGF-beta (Transforming Growth Factor Beta)
- bFGF (Basic Fibroblast Growth Factor)
- Interleukin-8
- Angiopoietin
- Angiogenin
- PDGF (Platelet Derived Growth Factor)
How do tumor cells induce angiogenesis
- Tumor cells can attract stromal cell types, which in turn can promote angiogenesis
- Tumor cells may release enzymes (like MMPs) which degrade ECM and the basement membrane
- Sometimes other angiogenic growth factors are present in ECM. When it is degraded those are released leading to additional angiogenesis.
- Tumor cells may attract local endothelial cells, but can also attract precursor cells from distant sites like the bone marrow.
Name some Anti-Angiogenic growth factors
Angiogenic Inhibitors
- Thrombospondin-1: ECM glycoprotein. Stops endothelial cells from proliferating
- Arrestin: fragment of collagen type IV
- Tumstatin: fragment of collagen type IV
- Angiostatin: fragment of plasminogen
- Endostatin: fragment of collagen XVIII
- Fibulin: fragment of basement membrane
- Endorepellin: fragment of perlecan
- TIMP-2: inhibitor of matrix metaloproteinase-2
What biological processes rely on angiogenesis?
- Embryonic development
- Implantation of the placenta
- Wound healing
- Disease processes
- Diabetic retinopathy, psoriasis, rheumatoid arthritis
- Tumorigenesis
What are the stages of the cell cycle? What are their relative lengths?
- S Phase (8-12 hours)
- M Phase (1 hour)
- G1+G2 (11-13 hours)
Interphase is G1+S+G2
Rapidly proliferating cells go through the cell cycle faster
Why are there checkpoint controls in the cell cycle?
Checkpoints provide an opportunity for a cell to determine if it is in a favorable condition to continue in the cell cycle. Is all the DNA replicated? Are all chromosomes attached to the spindle? Is the environment favorable?
Checkpoints allow for delays due to internal issues such as damaged DNA or poorly attached chromosomes
Checkpoints also allow for accumulation of microenvironmental signals such as growth factors indicating a general need to cell cycle progression in the tissue bed.
Where are checkpoints located in the cell cycle? What happens at each checkpoint?
(1) Before entering S phase from G1– Is the environment favorable? Checkpoint prevents S phase entry by stimulating p53 that activates transcription of the CKI called p21 that inhibits both G1/S-Cdk and S-Cdk
(2) Before entering M phase from G2– is all the DNA replicated? Checkpoint prevents M phase entry by stimulating kinases that inactivate the Cdc25 phosphatase needed for M-Cdk activation
(3) Before cytokinesis in M phase– are all the chromosomes attached to the spindle? Mad2 binds unattached kinetochores and blocks Cdc20-APC induced destruction of securin by sequestering Cdc20
How do checkpoints operate?
By NEGATIVE signals designed to stop the action
How were cell cycle control genes initially predicted and validated? (list experiments)
Genetic approach: Temperature sensitive yeat mutants– cell cycle control genes were predicted by G1/S blockade. Essential kinases are rate0limiting for cell cycle progression, and if these kinases are non functional, cell cycling stops at point where the kinases are needed.
Biochemical approach: Xenopus frog egg cytoplasm combined with the nuclei from a frog sperm and ATP– caused cell-free mitotic cycle, providing evidence for proteins in the cytoplasm driving mitosis and causing chromosomes to replicate and condense
Cell biology approach: Imaging replicated DNA in single cells or cell populations provides kinetic and spatial information about S phase activity– can see DNA replication occuring via an image, and can see relative amounts of DNA increasing during S phase
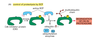
What are cyclins?
Cyclins are rate-limiting proteins that bind kinases (cyclin dependent kinases, or Cdk)
Cyclins undergo cycles of synthesis and degradation in each cycle
Cyclins and Cdk-activating kinase (CAK) are needed to activate Cdk
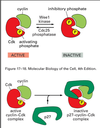
How is Cdk activity modulated?
(1) Inhibitory phosphorylation, such as Wee1 kinase with M-Cdk. The Wee1 kinase adds an extra phosphate which causes the inactivation of M-Cdk. Cdc25 phosphatase removes the extra phosphate to activate M-Cdk
(2) Cyclin-dependent kinase inhibitor (CKI), an inhibitory protein which binds to the active Cdk and caused an inactive CKI-Cdk complex, such as with G1/S Cdk and p27 CKI
(3) Ubiquitin ligase, reserves of cyclin:Cdk complexes can also be deployed or destroyed through ubiquitinization
–Anaphase Promoting Complex (APC) ligates ubiquitin to M-cyclin to end M phase
–SCF adds ubiquitin to p27 CKI to activate G1/S Cdk
Can the cytosol from an S phase nucleus trigger DNA replication in the G1 or G2 nucleus?
Only in G1 nucleus
What are the 4 phases of the cell cycle and what happens during each one?
G1: gathering nutrients and synthesizing RNA and proteins necessary for DNA synthesis
S: DNA replication
G2: cell growth and reorganization of organelles
M: mitosis
Describe the steps involved in replicating DNA before, during, and after S phase.
(1) Pre-replicative phase in G1: Cdc6 binds to the origin recognition complex (ORC) and MCM is recruited.
(2) S phase: S-Cdk phosphorylates Cdc6 causing its removal from the ORC and eventual degradation, MCM and phosphorylated ORC create replication fork.
(3) Upon the completion of DNA replication, M-Cdk phosphorylates MCM and Cdc6 creating a lack of the pre-replicative complex, so no more DNA can be replicated
Which cyclin-Cdk complex regulates G1 phase progression?
Cyclin D and Cdk4/6
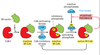
What happens at the DNA replication checkpoint?
- Unfinished replication forks send a negative signal to M-Cdk
- Negative signal activates a kinase that inhibits the Cdc25 phosphatase and allows Wee1 to keep M-Cdk in inactive state
- M-Cdk is very important in driving mitosis, so inhibiting it will allow for replication forks to finish
What does M-Cdk cause?
M-Cdk induces assembly of mitotic spindle, chromosome condensation, nuclear envelope breakdown, actin-myosin cytokinesis, and distribution of membranous organelles to daughter cells
How are sister chromatids separated?
M-Cdk causes two actions:
(1) Promotes Cdc20 to bind to inactive APC (anaphase promoting complex) to make active APC–> active APC causes the degradation of securin and activation of separase
(2) Mitotic spindle formation and attachment to the sister chromatids, separase cleaves cohesins
Now chromosomes can move toward the poles
How does the spindle-attachment checkpoint work?
Mitosis
- Sensor detects whether the sister chromatids are connected to spindle aparatus and moving to opposite pole
- If there is a failure in this, it will send a negative signal to block anaphase
- Mad2 binds unattached kinetochores and blocks Cdc20-APC induced destruction of securin by sequestering Cdc20
- M-Cdk and Polo kinase both release Cdc20 when all kinetochores become attached to the spindle, allowing for anaphase to proceed
How is M-Cdk degraded?
The APC-Cdc20 complex ubiquinates M-Cyclin
What happens to the Cdk activity in G1?
G1 phase = Cdk inactivity
CKIs accumulate to block G1/S-Cdk to prevent premature entry into S phase
APC creates new complex with Hct1 after Cdc20 removal– this assures full destruction of remaining M-cyclin and Cdc20 removal
How to cells move out of G1 and into S phase?
- G1-Cdk activation necessary– linkd to growth factor receptor activated kinases (MAP kinases such as Ras)
- Hct1-APC does not block G- or G1/S-cyclins
- G1-Cdk activates retinoblastoma protein (Rb) reducing its storage of E2F, a powerful transcriptional activator of genes needed for escape from G1 into S phase

How to mitogens affect exit from G1? What does Myc do?
- Mitogens bind to mitogen receptor, causing Ras/MAP kinase activation of gene regulatory protein and transcription of myc gene
- Myc protein causes:
- increased cyclin D–>G1-Cdk activation–> Rb phosphorylation
- increased p27 degradation–> G1/S-Cdk activation–> Rb phosphorylation
- Increased E2F synthesis
All of these cause increased E2F activity and entry into S phase
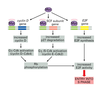
How does p53 work at the G1 checkpoint?
If DNA is ok, p53 is degraded
If there in an activating event (such as UV or X-rays), p53 is stabilized and binds to regulatory region of p21 gene, causing the creation of p21
p21 binds to G1/S-Cdk and S-Cdk preventing the cell’s progression into S phase
Describe the main features of Rb structure
The Rb gene codes for a large protein, retinoblastoma protein or Rb, (~150,000 daltons) that functions as a growth suppressor by binding to and sequestering a family of transcription factors known as E2F.
Phosphorylation of Rb by Cdk4/cyclin D and Cdk2/cyclin E results in hyperphosphorylation and release of the E2F transcription factors. Release and function of the E2F transcription factors allows cells to progress through the restriction point of the cell cycle.
What are three primary mechanisms that convert a proto-oncogene to an oncogene?
(1) Mutation
(2) Gene amplification (multiple copies of the gene)
(3) Chromosome rearragement (translocation or transposition, the gene is moved to a new locus under new promoter controls)
What is an oncogene?
A gene, which upon mutation, produces a protein that is over pexpressed or hyperactive resulting in excessive cell proliferation
Typically components of signal transduction pathways
Generally, a mutation if only one copy of a proto-oncogene is sufficient to disrupt growth regulation. Oncogenic mutations are thought to be dominant, and tumorigenesis results from the gain of function in the mutated oncogene.
What are possible consequences of proviral insertion?
When a retrovirus inserts DNA, it can cause retroviral transformation.
Proviral insertion may result in:
(1) Placement of long terminal repeat enhances sequences in proximity of a cellular proto-oncogene resulting in disruption of normal regulation of the proto-oncogene
(2) Insertion withing the coding sequence of a cellular proto-oncogene resulting in the production of an altered protein product that may have lost critical regulatory regions
(3) Insertion into regions of the gene that regulate proto-oncogene mRNA stability or protein stability, resulting in longer lived and/or greater amounts of protein
How can proto-oncogenes be activated by point mutations?
A mutation that inactivates a gene that has antiproliferative function can also lead to excessive proliferation.
In this case, the normal gene is referred to as a tumor-suppressor gene. In contrast to proto-oncogenes, usually both copies of a tumor-suppressor gene must be mutated or inactivated for an effect on growth regulation to become apparent. Tumorigenesis thus may also result from a loss of function of a tumor-suppressor gene.
What is retinoblastoma (Rb) protein?
Several roles:
(1) Functions as a growth suppressor by binding to and sequestering a family of transcription factors known as E2F. Regulates passage through G1/S restriction point of cell cycle. Phosphorylation of Rb by Cdk4/cyclin D and Cdk2/cyclin E results in hyperphosphorylation and release of the E2F transcription factors. E2F allows cells to progress through the restriction point of the cell cycle.
(2) Rb can capture SKP2/SCF and thus prevent the degradation of p27 CKI. The continued availability of p27 continues the inhibition of Cdks and keeps the cell in G1.
(3) Assists with the maintenance of chromatin structure during G1 and M phase
Mutations in Rb cause a wide variety of cancers.
What is Rb protein called the “master regulator”?
The restriction point monitored by Rb protein blocks passage through the remainder of the cell cycle
Passage through the restriction point normally allows cells to progress though the remainder of the cell cycle and divide once.
Other cell cycle checkpoints monitor cell cycle progression and, if activated, delay progression or repair or trigger apoptosis if damage is too great.
How does Myc modulate Rb phosphorylation?
Myc causes transcription of cyclin D gene, yielding increased cyclin D protein, causing G1-Cdk activation (cyclin D-Cdk4), which phosphorylates Rb
Myc increases transcription of SCF which degrades p27. Decreasing p27 allos for G1/S-Cdk (cyclin E-Cdk2) activation, which phosphorylates Rb
Describe the events in the cell after Rb phosphorylation.
Hyperphosphorylation of Rb protein (by G1-Cdk) inactivates Rb and allows for the release of E2F protein.
Active E2F protein increases S-phase transcription (which in turn activates more E2F protein. G1/S-cyclin (cyclin E) and S-cyclin (cyclin A) are transcriped. Newly activated G1/S-Cdk and S-Cdk positively reinforce S-phase transition by phsophorylating more Rb protein.
The cell exits G1 phase and enters S phase.
How does E2F function?
- Transcription factor
- Five members: E2F1-5
- Trans-activate cell cycle associated genes
- Form heteromeric complexes needed for induction of gene transcription
- Regulated by direct interaction with pocket proteins
How does the Rb pocket work?
When Rb is not phosphorylated, E2F sits inside the Rb pocket.
When Rb is phosphorylated, it causes Rb to change shape, and E2F can no longer fit in the pocket, and is thus released.
What are chromatin regulators?
Chromatin regulators are a variety of co-regulatory proteins that interact with Rb protein and facilitate conformation changes needed to release its negative control of E2F transcription factor activity.
Chromatin regulators may utilize the LXCXE pocket to allow for E2F dissociation.
What is an E2F independent mechanism for Rb-induced cell cycle arrest?
Rb can capture S-phase kinase associated protein 2 (SKP2/SCF) that adds ubituitin to the p27 cyclin-kinase inhibitor (CKI) needed for p27 turnover. p27 degradation is needed for progression through G1.
What are cohesins? And condensins?
Cohesins are multisubunit proteins that are deposited along the length of replicated chromatin to link sisters together.
Condensin proteins cause replicated chromatin to condense about 50-fold to produce visible chromosomes.
M-Ckd activates condensins which use ATP to drive coiling proces.
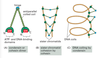
How do Rb mutations lead to genomic instability independent from cell-cycle progression?
In addition to the cell cycle restriction point functions, Rb protein also assists with the maintenance of chromatin structure during G1 and M phase
- Rb mutations can lead to defects in loading of condensin along prometaphase chromosomes and cohesin at the centromere.
- Condensin promotes condensation and cohesin holds the sister chromatids together
- Unregulated E2F causes high expression of MAD2 that prevents timely attachment of chromosomes to spindle microtubules.
- Lack of Rb and E2F excess results in tangled chromosomes and aneuploidy.