Genetics of antibodies and Tcell development Flashcards
19.08.01 Abbas(51-72) how it works(42-54), pharaoh(120-135)
affinity maturation
process of decreases Kd
avidity
measurment of the affinity of a single antigen-antibody bond
cross-reaction
antibodies produced against one antigen bind to another similar antigen
monoclonal antibodies
B cells that make one specific antibody
How is it possible to have a B cell reprtoire of 10^11?
germ line in-rearranged Ig, in non-B cells
somatic cell- each B cell undergoes somatic hypermutation
process that occurs during pr-b cell stage
- 12/23 rule
- RSS marks the recombination spot
- Rag joins regions and forms the loop and cuts the loop
- TdT adds nucleotides to fill in the gap
chains
- light chain
- combines VJ regions
- heavy
- combines DJ first
- then combines V with DJ
label and explain regions

- antibody L chain and H chain both have
- V region
- higher degree of variability in the N-terminal region
- C region
- limited variation
- V region
- antigen binding site
- formed by paired V regions
-
BCR=high affinity in Ab
- compared to lower affinity TCR
list the 5 Ab, chains, regions and domains
heavy chain
- C region
- alpha, delta, gamma
- C domain-4
- mu, epsilon
- extra domain is an elongated hing region
light chain
- isotypes
- kappa and lambda
- either is present in any individual antibody molecule
- no difference between the two
define the following
- isotype
- allotypes
- idiotypes
- isotypes
- antibody classes, five majore
- MDGEA
- difference lies in the heavy chain
- Mu, delta, gamma, epsilon or alpha
- light chain
- kappa or lamba
- antibody classes, five majore
- allotypes
- represent the genetically determined differences in antibodies between people
- example
- you and I both hav IgG, but unless we’re closely related, my IgG’s are very slightly different than yours
- used in paternity testing
- idiotypes
- antibodies of different specificity found within the same individual due to the diversity of the Ig V region
describe multivalence, with respect to the antigen, and its role in antibody attachment

what are two ways the Ab recognize epitopes?
l
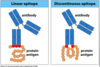
What confers epitope recognition and binding diversity?
Gentically hypervariable regions of antibodies
- framework regions
- regions that flank the hypervariable regions, these are much less variable
- hyper variability
- the differences in the aa sequence of the V domains between different antibody molecules are concentrated within particular locations in the gene.
- this is the V domain
- the differences in the aa sequence of the V domains between different antibody molecules are concentrated within particular locations in the gene.
- complementarity-determining regions (CDRs)
- another name for the hypervariable loops, since they provide the binding surface that is complementary to that of the antigen

describe the loci associated with Ig heavy vs light
light-two loci
- lambda loccus-c’some22
- kappa loccus-c’some 2
heavy
- one chain locus-c’some 14

start with a germ line DNA of heavy and light, and draw lines the final product after VDJ recombination. leave out lengthy mech parts

several steps are involved in rearrangment/recombination. Describe the recognition site
RSS- recombination signal sequences
- specal DNA motifs direct the siter of recombination in germ-line immunoglobulin DNA
- 12/23 rule
- RSS with 12bp spacer interacts only with RSS with 23bp spacer to ensure that gene segments are joined in the correct order

several steps are involved in rearrangment/recombination.
describe the joining step of the 12/23
RAG 1 and 2 are the cleaving enzyme
- only expressed in lymphocytes (B and T cells )
- bring the 12/23 spacers between D and J region
- cleave the DNA

What is the difference between combinational diversity and junctional diversity.
describe the junction components and resolving the junciton
combinational
- VDJ recombinations + light/heavy chain comombinations
junctdional diversity
- provides third CDR
- junctional flexibility-imprecision in the site of the join between gene segments
- P-nucleotides additions
- palindromes from loop cleavage and repair
- another enzyme cuts near the end of the RSS allowing nucleotides from the bottom (non-coding) strand to become part of the coding strand
- N-nucleotide additions
- terminal deoxy-nucleotidyl transferase (TDT) adds a random number of random nucleotides
resolving the junction
- b/c the number of disordered processes occurring, ther is a chance of generating “stop” codons orintroducing framshifts in the J region
- some “Joins” will be non-productive
- non-coding
- productive joins will generate increased diversity in the thir hypervariable region
- (CDR3)

describe the 3 CDRs
Complementarity-determining regions/hypervariable regions
- germ line contains varying sequences on
- CDR1 and 2
- junctional diversity is responsible for the increase in diversity at
- CDR3
allelic exclusion
- allelic exclusion
- process of shutting off the other allele after rearrangment of 1H and L chain. IF the first recombination works
- principle idea of specificity
- one H + one L =one specific
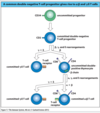
what is clonal selection?
what does it allow for?
clonal selection
- B cells are monospecific and for a given antigen only a subset of V cells wil produce antibodies to that antigen
- antigen specificity
- each b cell is specific for only one epitope of an antigen
- antibody repertoire/B cell reperetorre
- the numbe rof different antibodies/B cells recognizeing
describe the structures of the human Ig.
- variation in the C region of heavy chain determines the Ig Isotype (anitbody classes
- IgM-mu
- G-gamma
- D-delta
- A-alpha
- E-epsioln
- light chains have kappa or lambda, no difference in known

each b cell is specific for only one epitope, is an example of ?
antigen specificity
somatic recombination
aka: VDJ recombination
the process of DNA recombination by whic hthe functional genes encoding the variable regions of antigen receptors are formed during lymphocyte development.
- germline
- limited
- somatic recombination
- process occurs in developing B and T cells
- mediated by
- RAg 1 and RAG-2
2.
- RAg 1 and RAG-2
describe the ways B cells generate such diversity in the antibodies
- germ line
- hyper variable regions (CDR1&2)
- somatic recombination-gene rearrangment
- VDJ recombination
- 1.6x10^6 combinations
- junctional diversity (CDR3)
- VDJ recombination
- mRNA splicing-isotype
- after Ag exposure
- somatic hypermutation
- AID assisted isotype switching
Which type of Ig do naive B cells express? How?
naive B cells express both suface IgM and IgD
expression of two antibody isotypes simultaneously is acomplished by alternative mRNA splincing