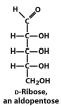
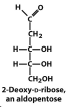
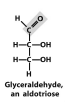
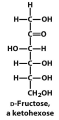
Exam III Flashcards
Michaelis Menton Equation
Define Vmax
- Vmax = kcat[E]o
- Vmax is the product of the catalytic rate constant (or turnover number) and the total enzyme concentration in the assay. Therefore, Vmax depends on the conditions of the assay. The important parameter is kcat, which is constant for a given reaction at standard conditions.
Define Km
- Km is the concentration of substrate at which Vo equals ½Vm
- Km = (k-1 + k2)/k1
- In order to maximize the biological efficiency of an enzyme, the Km of an enzyme is typically close the physiological substrate concentration.
- Higher Km indicates lower affinity for substrate.
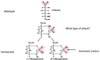
Lineweaver-Burke Plot
Significant points on the Lineweaver-Burke Plot
- The y-intercept of such a graph is equivalent to 1/Vmax
- The x-intercept of the graph represents −1/K<em>m</em>.
- The slope of the graph represents Km/Vmax.
If 10mg of pure carbonic anhydrase (29000 g/mol) catalyzes the hydration of 0.3 g of CO2 in 1 min at 37oC under optimal conditions, what is the turnover number, kcat, of carbonic anhydrase in units of min-1 and sec-1.
You use the equation: Vmax = kcat[E]o
To calculate Vmax: -d[CO2]/dt = 0.3g/min = 0.007 mol/min
To calculate [E]o: 10 micrograms / 29000 g/mol = 3.4 x 10-10 mol.
kcat = 0.007 mol/min / 3.4 x 10-10 mol.
- In a first order reaction a substrate is converted to product so that 87% of the substrate is converted in 7 min.
- Please calculate the first order rate constant for the reaction.
- Please calculate the half-life of the reaction-the time required for conversion of 50% of substrate to product.
A.) ln[A]/[A]o = -kt so ln[0.87]/[1.0] = -k(7 minutes) = 0.02 min-1
B) t1/2 = 0.693/k so t1/2 = 0.693/0.02 = 34.65 minutes.
Given a rate constant of 10.24 s-1 and the equilibrium constant for the dimerization process measured under the same conditions is 1.8x106M-1. What is the rate constant governing the dimer dissociation?
Keq = 1.8 x 106 M-1
ku = 10.24 s-1
Thus, kf = 1.8 x 10-7 s-1
First Order Reactions:
- Determine rate order constant
- Determine half-life
- To determine the rate constant, you use the equation –kt = ln[A]/[Ao]. You plot ln[A]/[Ao] vs time, the slope is equal to –k.
- The units of the rate constant are t-1 (time-1).
- To find the half-life: t1/2 = 0.693/k
Second Order Reactions:
- Determine the rate constant
- Determine the half-life
- To determine the rate constant, you use the equation 1/A = kt + 1/Ao. You plot 1/[A] vs time and the slope is equal to k and the y-intercept is equal to 1/Ao.
- The units of the rate constant are M-1t-1
- To find the half-life: t1/2 = 1/(kAo)
Reaction progress curve for an enzyme-catalyzed reaction.

Enzyme Catalysis Reaction
Experimental set-up: [S] >>> [E]o, initial rate conditions
- Set up several tubes, each containing a different substrate concentration [S]
- Add enzyme, E, to each tube. In all tubes [S] >>> [E]o
- Measure the product concentration versus time for each tube ([P] vs t)
- Plot [P] vs time at each [S]
- Determine the slope of each of these curves (d[P]/dt) and make sure that the initial region of the curve is being calculated so that the reverse reaction is not contributing.
- Plot initial rate, vo, versus [S]
- As [S] increases, Vo increases
- At relatively low [S], the Vo increases almost linearly with increases in [S]
- As [S] increases, Vo increases
Kinetic equation describing the reaction with microscopic rate constants
-
Km = (k-1 + k2)/k1
- If k2 is the rate limiting step, then Km approximates the equilibrium dissociation constant of the ES complex (k-1/k1)
- If k-1 is the rate limiting step, then Km approximates k2/k1
Steady-State Assumption
The steady state assumption is that at any single substrate concentration, the concentration of the enzyme-substrate complex initially rapidly increases to a level that depends on substrate concentration and remains constant during the time required to measure the initial rate.
Essentially, d[ES]/dt = 0.
Significance of kcat/km
- Different enzymes have similar efficiencies, but different Km and kcat values
- The best parameter for comparing activities of various enzymes or various substrates of the same enzyme is the Specificity Constant which is defined as the rate constant for conversion of E+S into E+P which is obtained from the first part of the vo vs [S] curve.
- The larger the parameter, the more efficient the enzyme. This parameter has the units of a second order reaction: M-1s-1
pH dependence of the rate with breakdown into effects on kcat and Km
- Find kcat vs pH
- The highest kcat will be at the optimal pH
- Find 1/Km vs pH
- When Km (binding of substrate and all intermediates), and when Km gets larger, the affinity for substrate decreases.
- Looking at a rate vs pH profile:
- Side Chain 1 is being deprotonated on the left side of the graph & Side Chain 2 is being deprotonated on the right side of the graph
- So this shows that the max activity is when Side Chain 1 is deprotonated and when Side Chain 2 is protonated.
- This shows that Side Chain 1 acts as a base and Side Chain 2 as an acid
- Side Chain 2 is also responsible for stabilizing the intermediate
Positive Allosteric Modulation
- Positive allosteric modulation (also known as allosteric activation) occurs when the binding of one ligand enhances the attraction between substrate molecules and other binding sites.
- An example is the binding of oxygen molecules to hemoglobin, where oxygen is effectively both the substrate and the effector. The allosteric, or “other”, site is the active site of an adjoining protein subunit. The binding of oxygen to one subunit induces a conformational change in that subunit that interacts with the remaining active sites to enhance their oxygen affinity.
Negative Allosteric Modulation
- Negative allosteric modulation (also known as allosteric inhibition) occurs when the binding of one ligand decreases the affinity for substrate at other active sites.
- For example, when 2,3-BPG binds to an allosteric site on hemoglobin, the affinity for oxygen of all subunits decreases. This is when a regulator is absent from the binding site.
Homotropic Allostery
- A homotropic allosteric modulator is a substrate for its target enzyme, as well as a regulatory molecule of the enzyme’s activity. It is typically an activator of the enzyme.
- For example, O2 is a homotropic allosteric modulator of hemoglobin.
Heterotropic Allostery
- A heterotropic allosteric modulator is a regulatory molecule that is not also the enzyme’s substrate. It may be either an activator or an inhibitor of the enzyme.
- For example, H+, CO2, and 2,3-bisphosphoglycerate are heterotropic allosteric modulators of hemoglobin
K-Type Regulation
Affects Km

V-Type Regulation
Affects Vmax.

Competitive Inhibition
This inhibition typically displays an unaffected Vmax, but a higher Km value. Affects “E + S” complex.