Exam 2: RNA Processing Flashcards
(55 cards)
What types of cells are we mainly talking about in this flashcard set?
EUKARYOTES! For our purposes, there is NO mRNA PROCESSING in prokaryotes. Prokaryotes do process rRNA and tRNA, but that is outside the scope of this flashcard set
How are final rRNA and tRNAs different from transcribed pre-rRNA and pre-tRNAs?
They are post-translationally modified by cleavege from precursors, other covalent modifications.
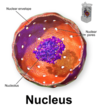
Where a pre-rRNAs cleaved to form functinoal RNAs?
In the nucleolus (shown in purple)
Further reading:
The nucleolus is the largest structure in the nucleus of eukaryotic cells, where it primarily serves as the site of ribosome synthesis and assembly. Nucleoli are made of proteins and RNA and form around specific chromosomal regions.
Three major components of the nucleolus are recognized: the fibrillar center (FC), the dense fibrillar component (DFC), and the granular component (GC).[2] The DFC consists of newly transcribed rRNA bound to ribosomal proteins, while the GC contains RNA bound to ribosomal proteins that are being assembled into immature ribosomes.
tRNAs are processed by?
Ribonuclease (commonly abbreviated RNase) is a type of nuclease that catalyzes the degradation of RNA into smaller components. Ribonucleases can be divided into endoribonucleases and exoribonucleases.
Made of RNA and protein, both parts functional, but RNA is the catalytic agent. Because of this, ribonucleases are a subclass of ribozymes
What type of macromolecule provides the catalytic ability of a ribozyme
RNA.
tRNA ribonucleases fall under this category, they contain both RNA and protein subunits. However, RNA contains the catalytic ability
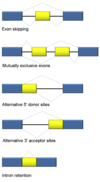
What is the difference between processing of mRNA in prokaryotes and eukaryotes?
mRNA in eukaryotes undergo many processing steps like 5’ capping, 3’ poly(A) tail, intron splicing (removal) before export from the nucleus. Prokaryotes do not undergo these modifications, infact in prokaryotes transcription and translation may occur simultaneously!
How many proteins does each mRNA code for?
Generally only one in eukaryoes, many in prokaryotes
Why do the ends of eukaryotic mRNAs need special processing (5’ cap, 3’ poly(A) tail)?
RNA polymerase in eukaryotes does not terminate as precisely as it does in prokaryotes
Why are 5’ caps added in eukaryotic mRNAs but not in prokaryotes?
Prokaryotes transcribe and translate simultaneously, their RNA need not be as stable because they don’t need the extra transport time to get out of the nucleus. Capping marks mRNA for export and prevents accidental degredation by exonucelases. It also is required for translation and can be used to control expression.
Structure of the 5’ cap and funtion
The 5’ cap is a guanine nucleotide connected to mRNA via an unusual 5′ to 5′ triphosphate linkage. This makes it appear chemically similar to the 3′ end of an RNA molecule (the 5′ carbon of the cap ribose is bonded, and the 3′ unbonded), providing significant resistance to 5′ exonucleases (prevents degredation).
Further reading:
In eukaryotes, the 5′ cap (cap-0), found on the 5′ end of an mRNA molecule, consists of a guanine nucleotide connected to mRNA via an unusual 5′ to 5′ triphosphate linkage. This guanosine is methylated on the 7 position directly after capping in vivoby a methyltransferase. It is referred to as a 7-methylguanylate cap, abbreviated m7G.
In multicellular eukaryotes and some viruses, further modifications exist, including the methylation of the 2′ hydroxy-groups of the first 2 ribose sugars of the 5′ end of the mRNA. cap-1 has a methylated 2’-hydroxy group on the first ribose sugar, while cap-2 has methylated 2’-hydroxy groups on the first two ribose sugars, shown on the right. The 5′ cap is chemically similar to the 3′ end of an RNA molecule (the 5′ carbon of the cap ribose is bonded, and the 3′ unbonded). This provides significant resistance to 5′ exonucleases.
Nuclear export of RNA is regulated by the cap binding complex (CBC), which binds exclusively to capped RNA. The CBC is then recognized by the nuclear pore complex and exported. Once in the cytoplasm after the pioneer round of translation, the CBC is replaced by the translation factors eIF-4E and eIF-4G. This complex is then recognized by other translation initiation machinery including the ribosome.
Capping prevents 5′ degradation in two ways. First, degradation of the mRNA by 5′ exonucleases is prevented (as mentioned above) by functionally looking like a 3′ end. Second, the CBC and eIF-4E/eIF-4G block the access of decapping enzymes to the cap. This increases the half-life of the mRNA, essential in eukaryotes as the export and translation processes take significant time.
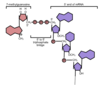
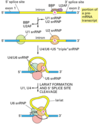
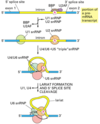
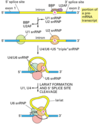
Describe the process of capping a eukaryotic mRNA
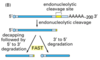
First, a phosphatase removes one phosphate from the very 5’ end (the 5’ end was added with a NTP (nucelotide triphosphate, and so has three phosphates at the very end. Removing one leaves two phosphates attatched to the 5’ end, now a NDP).
Next, guanylyl trasferase adds GMP in an unusual 5’-5’ triphosphate linkage (this addition is not the same as a regular addition, it is added in the 5’ to 3’ direction, which RNAPol II is not capable of. Notice it is catlysed by a different protein, guanylyl trasferase. GMP is a mono phosphate, so added to the NDP makes a triphosphate 5’-5’ linkage).
The G is then modified by methyl transferasese, which add one or more methyl groups (see below for further reading)
The 7-nitrogen of guanine is methylated by mRNA (guanine-N7-)-methyltransferase, with S-adenosyl-L-methionine being demethylated to produce S-adenosyl-L-homocysteine, resulting in 5’(m7Gp)(ppN)[pN]n (cap-0);
Cap-adjacent modifications can occur, normally to the first and second nucleotides, producing up to 5’(m7Gp)(ppN*)(pN*)[pN]n (cap-1 and cap-2);[7][8]
If the nearest cap-adjacent nucleotide is 2’-O-ribose methyl-adenosine (i.e. 5’(m7Gp)(ppAm)[pN]n), it can be further methylated at the N6 methyl position to form N6-methyladenosine, resulting in 5’(m7Gp)(ppm6Am)[pN]n.
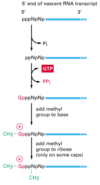
When are mRNAs capped? How do the capping factors find the mRNAs?
Caps are put on pol II transcripts very soon after initiation. When the nascent pre-mRNA is about 25 nucleotides long, the process begins.
The capping enzymes ride along on the phosphorylated tail (CTD: carboxy terminal domain) of the RNA pol II large subunit.
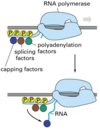
endonuclease
Endonucleases are enzymes that cleave the phosphodiester bond within a polynucleotide chain. Some, such as Deoxyribonuclease I, cut DNA relatively nonspecifically (without regard to sequence), while many, typically called restriction endonucleases or restriction enzymes, cleave only at very specific nucleotide sequences.
exonuclease
Exonucleases are enzymes that work by cleaving nucleotides one at a time from the end (exo) of a polynucleotide chain. A hydrolyzing reaction that breaks phosphodiester bonds at either the 3’ or the 5’ end occurs.
Its close relative is the endonuclease, which cleaves phosphodiester bonds in the middle (endo) of a polynucleotide chain.
Eukaryotes and prokaryotes have three types of exonucleases involved in the normal turnover of mRNA: 5’ to 3’ exonuclease, which is a dependent decapping protein; 3’ to 5’ exonuclease, an independent protein; and poly(A)-specific 3’ to 5’ exonuclease
what type of nuclease would you use for decapping a mRNA (removing either a 5’ cap or 3’ poly(A) tail)?
Exonucleases are enzymes that work by cleaving nucleotides one at a time from the end (exo) of a polynucleotide chain.
Eukaryotes and prokaryotes have three types of exonucleases involved in the normal turnover of mRNA: 5’ to 3’ exonuclease, which is a dependent decapping protein; 3’ to 5’ exonuclease, an independent protein; and poly(A)-specific 3’ to 5’ exonuclease.
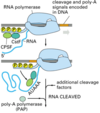
Where is the poly(A) tail added?
To specific sequences near the 3’ end of the pre-mRNA that direct cleavage and polyadenylation

what does poly(A) stand for? basically what does it mean?
polyadenylation
the addition of many repeating adenine residues

Where can poly(A) factors be found?
The polyadenylation factors ride along on the phosphorylated tail (CTD: carboxy terminal domain) of the RNA pol II large subunit.
What are the roles of the cap and the poly(A) tail?
The cap protects the 5’ end from 5’ to 3’ exonucleases. The poly(A) tail protects the 3’ end from 3’ to 5’ exonucleases.
Thus, both are important in regulating the half-life of the mRNA (eukaryotes need more time than prokaryotes, they must move the mRNA out of the nucleus)
What can happen to the poly(A) tail in the cytoplasm?
For some mRNAs, the poly(A) tail gradually shortens during its life in the cytoplasm. When it reaches a critical length, a de- capping enzyme is activated that removes the cap. The mRNA is then rapidly degraded in a 5’ to 3’ direction.

How might an endonuclease shorten the life of a eukaryotic mRNA in cytoplasm?
For some mRNAs, the poly(A) tail gradually shortens during its life in the cytoplasm. The endonuclease may remove a portion of the AAA tail, causing the rest to shorten to the critical length much faster. When it reaches a critical length, a de- capping enzyme is activated that removes the cap. The mRNA is then rapidly degraded in a 5’ to 3’ direction
What is the difference in stability between dsDNA, dsRNA, and DNA:RNA hybrids?
RNA:RNA > RNA:DNA > DNA:DNA
tl;dr: It’s all about the helix! The addition of the 2’ hydroxl does less to effect stability but sterically hinders the types of helix RNA can stabily adopt.
RNA is limited to B form, while DNA can adopt B, A or Z form.
RNA is more stable than DNA
(further reading)
One major difference between RNA and DNA is the detailed shape of the double-helix, A-form for RNA and predominantly B-form for DNA. RNA has never been observed to take on a B double-helix; the presence of the 2’-OH almost exclusively locks the ribose into a 3’-endo chair conformation, eliminating the possibility of a stable B-helix. However, the deoxyribose sugar may alternate between 2’-endo and 3’-endo conformations, allowing DNA to switch between B-form and A-form under the right circumstances. Note that hybrids of DNA:RNA (one strand of each in a double-helix) adopt an A-form conformation.
∆G = ∆H - ∆S(T)
In total, the single-strand to double-strand transition for both DNA and RNA is enthalpically (H) favors the helix and entropically (S) favors single-stranded conformation. For RNA, ∆H ~ 40 kJ mol-1/base pair and ∆S ~ 105 J K-1 mol-1/base pair (note the entropy is a function of temperature). For DNA, ∆H ~ 35 kJ mol-1/base pair and ∆S ~ 90 J K-1 mol-1/base pair. These are VERY large and OPPOSITE driving forces
In terms of the free energy, the balance of these interactions, we observed a higher melting temperature of RNA relative to the same sequence in DNA under normal conditions. The dominant source of this slightly higher energy for RNA is generally attributed to modestly better base-stacking energy in the A-form conformation. The precise nature of the molecular driving forces remain an active area of research. Experimentally, one observes very little difference in thermostability between RNA:DNA double-helix compared to an all RNA double-helix, consistent with the theory that the source of thermostability is due largely the result of A-form versus B-form conformational differences, not strictly differences in ribose versus deoxyribose chemistry.

How did biologists first determine mRNA and genes are not 1:1 matches in eukaryotes?
By studing a gene from a human virus because:
- human viral genes are expressed in human cells
- the viral genome is simple to purify
- the viral mRNAs are expressed at high levels and easy to purify
Purify DNA from Adenovirus virions, mRNA from infected cells.
Study by R-looping: hybridize DNA and mRNA. The DNA:RNA hybrid is stronger; the RNA will hybridize to the template strand of the DNA and displace the sense strand. Visualize using electron microscopy.
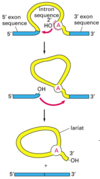
Results is mRNA is bound to complimentary areas (exons) and large loops exist in DNA where RNA is not bound (introns).

intron
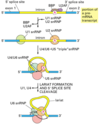
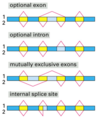
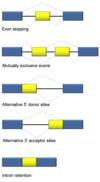
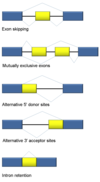
Introns are found in all types of RNAs (mRNA, rRNA, tRNA etc) in all types of organisms.
In splicing, the precursor RNA is made into a mature RNA and the intron is released.
No ATP needed for the splicing reaction because no new phosphodiester bonds are made.