Exam 1: Molecules and Cells/Water, pH, and Buffers/Nucleic Acids/Nucleic Acid Structure Flashcards
hold atoms together to form molecules.
Covalent bonds
maintain biological structure and determine molecular interactions.
Noncovalent bonds (or weak chemical forces)
Types of noncovalent bonds:
- Van der Waals Interactions: result of electron cloud fluctuations that allow attractions to occur between positively charged nuclei and electrons of nearby atoms
- Hydrogen Bonds: form between a hydrogen atom, covalently bonded to an oxygen or nitrogen, and a second oxygen or nitrogen
- Ionic Interactions: result of attractive forces between oppositely charged groups, such as negative carboxyl groups and positive amino groups
- Hydrophobic Interactions: due to the strong tendency of water to exclude nonpolar groups or molecules, because water molecules prefer the stronger interactions between water molecules
Compared to similar liquids, water has a higher:
1.
2.
3.
4.
- Boiling point
- Melting point
- Heat of vaporization
- Surface tension
The unusual properties of water are due to its molecular structure:
a) Water is a polar molecule: The electronegative O atom and the two H atoms form a dipole that makes the molecule polar.
b) Hydrogen bonds are formed by interactions between the polar water molecules: H2O molecules in liquid water interact with an average of 4.4 other molecules to form a random, H-bonded network. These intermolecular attractions are responsible for the high boiling point, etc. of water.

The H bonds in ice form a rigid structure which holds the water molecules apart.
Because of its structure, it is sometimes called ice, or structure
hexagonal, open lattice

The average lifetime of an H-bond between two H2O molecules is picoseconds (1 psec = 10-12 sec).
9.5 picoseconds
Because water is a polar molecule, it is an excellent for many substances including
solvent,
ionic substances such as salts.
H2O molecules form around ions.
hydration shells
Negative partial charges on oxygen will be attracted to the positive charge of Na+, while the partial positive charges on the hydrogens will be attracted to the negative charge of Cl-.

Water Has a High Dielectric Constant
The ability of water to surround ions in dipole interactions and diminish their attraction for one another is a measure of its dielectric constant, D
water molecules form around hydrophobic solutes.
clathrate structures

Water Forms , which accounts for its excellent solvent properties.
H Bonds with Polar Solutes
Water molecules produce with molecules. Examples are fatty acids with a polar group (carboxyl group) at the end of a long nonpolar chain.
micelles
amphiphilic: means that there a two chemical natures within one molecule

two nonpolar tails instead of one results in a very different structure than amphiphilic molecules containing just one nonpolar tail – a instead of a micelle.
bilayer
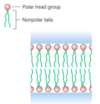
the lipid bilayer helps to maintain the different concentrations of ions on the inside and outside of the membrane.

What are colligative properties?
Give examples for water
Properties that only depend on the number of solute particles per unit volume of solvent and not on the chemical nature of the solute.
EX:
- freezing point depression
- boiling point elevation
- vapor pressure lowering
osmotic pressure effects
They only depend on the amount of molecules mixed into the water.
Mixing certain starches or proteins with water can lower the freezing point of water than 32 degrees, for example. All these molecules are doing is interfering with the ability of the water molecules to form a crystal hexagonal structure, so you have to slow down the water molecules even more to get them to freeze.
Ions in solution, such as the Na+ and Cl- ions of NaCl, interact with H2O molecules to form:
a) clathrate structures
b) hexagonal ice
c) micelles
d) osmotic pressure
e) hydration shells
e) hydration shells
The structure of water in the form of normal ice can be described as “hexagonal ice”. This is because:
a) ice crystals always have six sides
b) hydration shells consist of six H2O molecules
c) the minimum number of water molecules around any closed path of H-bonded molecules is six
d) clathrate structures of water molecules are composed of six H2O molecules e) ice floats on liquid water
c) the minimum number of water molecules around any closed path of H-bonded molecules is six
If the initial pH of a solution is pH 8.0 and acid is added to increase the hydrogen ion concentration 100-fold, the final pH of the solution will be:
a) pH 8.0
b) pH 6.0
c) pH 7.0
d) pH 1.0
e) none of the above
b) pH 6.0
In the phosphate buffer system, which buffers the intracellular fluid of cells, excess acidity [H+] is neutralized to form H2PO4- by reaction with:
a) OH-
b) NaH2PO4
c) H2O
d) HPO42-
e) H2CO3
d) HPO42-
Which of the following nucleic acid bases are pyrimidines only?
a) guanine, adenine
b) cytosine, uracil, thymine
c) guanine, cytosine, adenine, thymine
d) guanine, adenine, thymine
e) guanine, cytosine, adenine, uracil
b) cytosine, uracil, thymine
Which bases are the pyrimidines commonly found in RNA?
a) guanine, cytosine
b) guanine, adenine
c) cytosine, uracil
d) adenine, uracil
e) adenine, thymine
c) cytosine, uracil
Which of the following is a nucleoside?
a) adenine
b) adenosine
c) adenosine 5’-monophosphate
d) adenylic acid
e) 3’, 5’-cyclic AMP
b) adenosine
All of the following nucleotides are substrates for the synthesis of RNA or DNA in cells except:
a) thymidine 5’-triphosphate
b) uridine 5’-triphosphate
c) deoxycytidine 5’-triphosphate
d) deoxyadenosine 5’-triphosphate
e) guanosine 5’-triphosphate
a) thymidine 5’-triphosphate
- I think this should be deoxythymidine. Thymine is only found in DNA; therefore it would have to be deoxy?










