DNA Replication and General Features Flashcards
(20 cards)
Outline the different models of replication that were possible before being experimentally determined.
Watson and Crick proposed that DNA replicated in a conservative manner, however this didn’t rule out the possibilities of conservative and dispersive replication.
- In the semi-conservative model, the two parental strands separate and each makes a copy of itself. After one round of replication, the two daughter molecules each comprises one old and one new strand. Note that after two rounds, two of the DNA molecules consist only of new material, while the other two contain one old and one new strand.
- In the conservative model, the parental molecule directs synthesis of an entirely new double-stranded molecule, such that after one round of replication, one molecule is conserved as two old strands. This is repeated in the second round.
- In the dispersive model, material in the two parental strands is distributed more or less randomly between two daughter molecules. In the model shown here, old material is distributed symmetrically between the two daughters molecules. Other distributions are possible.

How was the model of replication determined?
Method of replication was determined experimentally by Meselson and Stahl in 1958. They used an ultracentrifuge which could separate molecules based on very slight densities. In their experiment they used 15N to alter the density of DNA and grew E. coli in medium using 15N only as the only source of nitrogen, and then in 14N. The DNA is centrifuged in a CsCl solution, and the more dense molecules go towards the bottom of the tube (CsCl gradient brackets the N density). At equilibrium the ‘light’ 14N-containing strands will separate from the ‘heavy’ 15N-containing strands into two distinct zones. Samples are taken at intervals and centrifugation is carried out.
- Round 1: each duplex has one heavy and one light chain, so all daughter molecules have an intermediate density
- Round 2: one duplex is intermediate density, one it made from two ‘light’ chains so is ‘light’. 50/50 ratio.
- Subsequent rounds: the proportion of light chains increases as new 14N is used to synthesise new strands.
This distribution was what was expected of semi-conservative replication.

What type of centrifugation was used in the Meselson-Stahl experiment?
Equilibrium density-gradient centrifugation
How was it determined in the Meselson-Stahl experiment that the model for DNA replication was semi-conservative, and not dispersive?
After one generation, DNA is “Half-heavy”. This is consistent with both semi-conservative and dispersive models. After ~ two generations, DNA is of two types: “Light” and “Half-heavy”. This is consistent with only the semi-conservative model.

Give a brief overview of bacterial initiation of replication.
Prokaryotic chromosomes are usually circular, so there are no free ends. Circular structures include chromosomes, plasmids, and mitochondrial DNA. After initiation two replication forks proceed in opposite directions, a complication being that completion can give rise to two linked chromosomes (catenated chromosomes), which need specific enzymes to separate them.
The E. coli genome is replicated bidirectionally from the origin, OriC, until it reaches the ter (termination) site.

Outline the role of DNA methylation in the regulation of initiation in E. coli.
- Before DNA replication, GATC sequences throughout the E. coli genome are methylated on both strands (“fully” methylated).
- DNA replication converts these sites to the hemimethylated state (only one strand of the DNA is methylated).
- Hemimethylated GATC sequences are rapidly bound by SeqA.
- Bound SeqA protein inhibits the full methylation of these sequences and the binding of oriC by DnaA protein.
- When SeqA infrequently disassociates from the GATC sites, the sequences can become fully methylated by Dam DNA methyltransferase, preventing rebinding by SeqA.
- When the GATC sites become fully methylated, DnaA can bind the 9-mer sequences and direct a new round of replication from the daughter oriC replicators.
This slows the rate at which oriC is initiated, and therefore stops uncontrolled division.
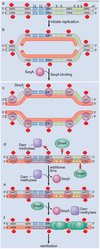
By what method do circular chromosomes replicate?
Bidirecctional replication
What is the origin of E. coli?
OriC
- 245bp long
- Contains 11 GATC methylation sites
- An AT-rich region
- DnaA boxes
Define origin of replication.
A sequence of DNA at which replication is initiated.
Define replication fork.
The point at which strands of parental duplex DNA are separated so that replication can proceed. A complex of proteins including DNA polymerase is found there.
Define bidirectional replication.
A system in which an origin generates two replication forks that proceed away from the origin in opposite directions.
How is replication initiated in E. coli?
DNA replication is initiated by the binding of DnaA proteins to the DnaA box sequences. 10-20 monomers of DnaA bind to the 9-mer repeats, and the DNA coils around this protein complex forming a protein core. This association of proteins leads to the separation of DNA strands. Additional proteins, DnaB/DnaC, joins the complex forming replication forks.

What six proteins form a complex when initiating replication in E.coli?
Initiation of replication at oriC starts with the formation of a complex that ultimately requires 6 proteins: DnaA, DnaB, DnaC, HU, Gyrase, and SSB.
- DnaA binds ATP and initiates replication when in a DnaA-ATP complex bound to fully-methylated oriC DNA. Opening the DNA requires binding 9-mer and 13-mer, which twists the DNA to open the DNA at AT-rich sites (13-mer sites).
- DnaB is helicase. Recruited in a complex with DnaC (chaperone protein) as hexamers.
- Gyrase is a type II topoisomerase.
- SSB stabilises ssDNA.
- HU is a general DNA-binding protein in E. coli.
Define replicon.
A unit of the genome in which DNA is replicated. Each contains an origin for initiation of replication.
Define ARS.
ARS = autonomously replicating sequence.
An origin for replication in yeast. The common feature among different examples of these sequences is a conserved 11 bp sequence called the A domain/ACS.

What is the origin of replication in S. cerevisiae?
Saccharomyces cerevisiae: In 1980, R. Davis and co-workers discovered that Autonomously replicating Sequence - ARS1 and ARS307, allow plasmids to replicate autonomously. Usually short (about 200bp) but also contains repeat elements. Eukaryotic chromosomes contain many replicons which can replicate independently, a sequence like this in yeast is an ARS. The yeast ARS element consists of an A-T-rich region, the consensus sequence being called ACS (ARS consensus sequence). Mutations in ACS and adjacent B elements can impact on origin function.
There are an estimated 400 origins of replication in the yeast genome.

What proteins bind to the origin of replication in S. cerevisiae (ARS)?
In 1992, Bell and Stillman isolated a yeast protein that binds ARS. This protein is called origin of recognition complex (ORC). The ORC is a complex of proteins that binds to the A and B1 elements. DNA sequences at the replication origins in Higher Eukaryotes, including humans, vary and have been much harder to isolate. Evidence suggests that the origin of replication in mammalian cells is similar to yeast type origins.
In other eukaryotes, all the origins form a base for assembly of a group of proteins known collectively as the pre-replication complex (pre-RC):
- First, the origin DNA is bound by ORC complex (six different protein subunits) which, with help from two additional cell division cycle proteins (Cdc6 and Cdt1), load the mini chromosome maintenance (or MCM) protein complex.
- Once assembled, this complex of proteins indicates that the replication origin is ready for activation. This process by which pre-RC is assembled is called licensing.

Where does replication take place in eukaryotes?
Replication factories.
Outline the differences in replication speed in humans and E.coli.
-
E.coli: DNA replication takes about 40 minutes
- Circular chromosome = 4.4 x106bp
- Replication from one origin by 2 forks (bi-directional) 50,000 basepairs/min
-
Human: the ~ 3 billion base pair genome takes 8hrs to replicate
- Between 10,000 – 100,000 replicons
- Replication fork speed of ~ 2000 basepairs/min


