DNA Repair Learning goals Flashcards
Why should we care about DNA repair?
- As the carrier of genetic information, DNA is the only macromolecule that is repaired. All others are replaced.
- DNA is continually exposed to endogenous and exogenous sources of damage, necessitating the activity of multiple repair pathways to maintain genomic integrity.
- DNA repair pathway alterations are cancer drivers.
- Tumor DNA repair pathway alterations are biomarkers as well as therapeutic targets.
- Tumor DNA repair deficiencies and mutation load can predict response to immunotherapy.

Types of DNA damage

Damage to base: hydrolysis, alkylation, UV

Damage to base: oxidative damage

Base deamination can cause point mutation

Base alkylation can cause point mutation

Changes in DNA structure can interfere with replication and transcription

Guanosine-BPDE adduct causes insertion of A opposite of G by DNA polymerase, leading to G->T mutation.
Types of DNA Repair
•Direct reversal of damaged bases
- Reversal of a specific type of single-stranded DNA break by DNA ligase.
- Reversal of UV-caused base damage (T-T T-C dimers) by photolyase.
- Reversal of base alkylation by O6-meG methyltransferase (MGMT).
•Excision of damaged, mispaired, or incorrect bases
- Base excision repair (BER)
repairs base damages that do not distort the DNA.
- Nucleotide excision repair (NER)
repairs base damages that distort the DNA.
- Mismatch repair (MMR)
removes misincorporated nucleotides during DNA replication.
•Tolerance/Bypass of base damage (trans-lesion synthesis)
•Strand break repair of damage to the DNA backbone
- Single-strand break repair (SSBR)
- Double-strand break repair (DSBR)
Homologous recombination (HR)
Nonhomologous end-joining (NHEJ)
Removal of a methyl group from
O6-methylguanine by methyltransferase (MGMT)

- MGMT (O6-methylguanine methyltransferase) is evolutionarily conserved and a classical example of “direct reversal” type of DNA repair.
- Tumor-associated mutations in MGMT reduces its DNA repair activity.
- MGMT is silenced via promoter methylation in ~45% of human glioblastomas.
Excision Repair: three types
Three types - all three repair machineries take advantage of the double-stranded nature of the DNA molecule to copy the correct information from the intact strand of DNA to the damaged strand.
1- Base excision repair (BER)
- repairs base damages that do not distort the DNA.
- uses specific glycosylases to remove the damaged base.
2- Nucleotide excision repair (NER)
- repairs base damages that distort the DNA.
- removes an oligonucleotide that contains the damaged base.
3- Mismatch repair (MMR)
- removes misincorporated nucleotides during DNA replication.
- distinguishes between the template strand and the new strand.
Steps common to all three excision repair mechanisms
- Recognition of the damaged/mismatched nucleotide.
- Endonuclease-mediated cutting of the phosphodiester backbone flanking the damaged/mismatched nucleotide.
- Nuclease-mediated removal of the DNA fragment containing the damaged/mismatched nucleotide.
- DNA polymerase-mediated synthesis of the missing nucleotides by copying nucleotide sequence from the intact DNA strand.
- DNA ligase-mediated sealing of the remaining nick in the phosphodiester backbone.
Base Excision Repair (BER) removes base damage
that doesn’t distort the DNA duplex
Base Excision Repair is initiated by glycosylase

Limited to only certain bases and mispairing.
Nucleotide Excision Repair (NER) removes damages that distort the DNA structure and block polymerase function
- A little more flexible than base excision repair.
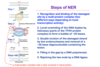
Two ways NER machineries recognize damage

Human diseases caused by defective NER

Mismatch Repair (MMR) corrects errors made by
DNA polymerase during replication
- S-dimers come to site before the L-dimers
- Requires helicases because it cuts long pieces of strand
How does it tell which is the template?
Due to absence of methylation in new strand
Image is from E.coli

How does MMR recognize
which is the right DNA strand to repair? In eurkaryotes

Nascent lagging strand is marked by transient 5’ DNA ends of short discontinuous Okazaki fragments.
Nascent leading strand is marked by transient presence of ***ribonucleotides (1 rNMP/1250 dNMP), which is processed into nicks by RNase H2 (trimer enzyme).
Mutations in MMR genes cause HNPCC
HNPCC is one of the most common inherited cancer-susceptibility
syndromes and accounts for ~5% of all cases of colon cancers.
(hereditary non-polyposis colorectal cancer: Lynch Syndrome)

Unique to each type of excision repair
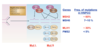
DNA damage tolerance/bypass:
trans-lesion synthesis
- Is used when cells have too much DNA damage for the “error-proof” repair machineries (NER, BER, MMR) to handle, especially replication-blocking lesions.
- As a last resort, cells employ “bypass” or “error-prone” DNA polymerases with loosened specificity to enable replication to continue through damaged template strand.
- The bypass polymerases lack the proofreading 3’-to-5’ exonuclease activity and have error rates 100-10,000 fold higher than those of the polymerases used in normal DNA replication.
Bypass DNA polymerases in translesion synthesis

Repair of double-strand breaks (DSBs)

Single-strand DNA breaks: causes and consequences

Reversible poly(ADP-ribosylation) of proteins near a single-strand break sites facilitates DNA repair

The DNA Damage Response: pausing the cell cycle for repair

DDR defects, mutation load and sensitivity to cancer immunotherapy
Tumors cells can present proteins in their surface that bind to T cells and prevent their activation.
For example the receptor PD-1 in T-cells bind to PD-L1 from cancer cells and turns off its tumor suppressing activity.

DNA Repair sheat cheet
A different schematic of the repair pathways.
Emphasis:
Direct reversal is always accurate
NER, MMR and BER are accurate
HR is mostly accurate
TLS is often inaccurate
NHEJ is often inaccurate



