Diagnostic and Typing Methods Flashcards
bacteria associated with periodontal disease (4)
- Porphyromonas gingivalis
- Actinobacillus actinomycetemcomitans
- Prevotella intermedia
- Bacteroides forsythus
bacteria associated with dental caries
sterptococcus mutans
bactaeria associated with root canal infections (endodontic)
- Porphyromonas endodontalis
- Fusobacterium nucleatum
2 classes of bacteria detection methods
microbiological culture
molecular biological
microbiological culture involves
- Culture on suitable agar medium
- Isolate bacteria
- Identify by characterisation of enzyme activities, sugar fermentation tests
traditional way to identify bacteria within clinical specimens.
molecualr biological
- DNA probes
- Polymerase chain reaction (PCR) very sensitive
microbiological culture methods
- Vortex mix sample for 30 seconds
- Serial dilutions (to 10-6) in FAB
- Spiral plate to agar media:
- Fastidious Anaerobe Agar (FAA) + 7.5% v/v defibrinated horse blood - Grow well
- As a) but supplemented with vancomycin (selective agent for Gram-negative anaerobes), mant oral diseases are caused by gram – anaerobes
- Incubate anaerobically for 10 days
- Obtain total bacterial counts
why need to serial dilute before plating
If plate the undiluted mixed sample there would be so many bacteria that we would not be able to discern individual colonies
black colonies on Fastidious Anaerobe Agar plat

varierty of different bacterial colonies of different shapes and sizes and colour
- black pigmented colonies which generally belong to the genera Porphyromonas and Prevotella,
- Associated with oral diseases such as periodontal disease
black colonies associated with
- black pigmented colonies which generally belong to the genera Porphyromonas and Prevotella,
- Associated with oral diseases such as periodontal disease
what to do after growing variety of colonies
Individual bacterial colonies on the surface of the agar plate can then be picked off (selected) and grown in pure culture
process of selecting particular colony to grow individually
subculture it
get pure colony of that particular species
Can see all individual colonies of that particular black pigmented species, can see that black pigment is produced, seeping out from the colonies into the agar surrounding it.

what to do once have a pure culture
ID
biochemical identification
3 methods of biochemical identification
- anaerobes noted by their sensitivity to metronidaxole disc (5ug/disc)
- gram stain
- rapid API 32 A:enzymatic activites, sugar fermentation (commercially available kits)
gram + ->
violet
gram - –>
pink
how use information from rapid API 32A
identify bacteria by creating an enzymatic activity and sugar fermentation profile and comparing those activities to those held centrally in a database
identify the bacteria on the basis of which enzymatic activities they possess and which sugars they can ferment to produce acid from.
how to ID Anaerobes by their sensitivity to metronidazole disc (5μg/disc)
Fastidious Anaerobe Agar plate that contains a pure culture of a particular bacterial species.
Metronidazole disc
- zone of clearing around the metronidazole disc.
- Bacteria around the disc have died.
- tells us that this particular bacterial species that we’re growing on the surface of this agar plate is a Gram-negative anaerobe.
- Bacteria around the disc have died.

ID from 2 samples of results following enzymatic activity and sugar fermentation biochemical identification tests.
rapid API 32A

2 strips which have several cupules containing sugars or enzyme substrates.
- In the cupules containing yellow fluid, a variety of sugars have been fermented by the bacterial suspension that was placed into that cupule, thereby producing acid.
build up a biochemical profile of the bacteria that we are testing.
Creating a profile in this way based upon their enzymatic activities and sugar fermentation abilities, allows us to confirm the identify of bacterial isolates by comparing their biochemical profiles to those for known bacterial species which are contained within a central database.

advantage of culture methods
- Yields bacterial isolates for future testing and study eg. antibiotic sensitivities
5 disadvantages of culture methods
Requires viable cells
- Often bacteria causing infections will die off
Insensitive (usually need 105-106 cells)
Only small numbers of samples can be analysed at once – as labour intensive
Inconclusive results major limitation
- Biochemical identification methods are not hugely accurate for all species
- Ideally want 100% certainty (or at least 95% certainty) that a particular bacterial species you’ve isolated is matching with another species in the database to that level.
- Unfortunately, the biochemical identification methods can give us a match with bacteria in the database much lower than 95%, quite often 50 or 60% and that is inconclusive with regard to its identification. .
Labour-intensive and expensive
DNA Probes
basics of working
Segments of DNA that have been labelled with chemoluminescent, fluorescent or radioactive agents
3 types of DNA probes
- Whole genomic (entire genome)
- Cloned gene
- Oligonucleotide (20-50 bases) will target particular bacterial gene
DNA probes Vs culture
more sensitive
103 cells
first stage in DNA probe
prepare the probe for the sample
how to prepare DNA probe for sample
The probe is a DNA double-stranded molecule that must be pull apart so it’s heat denatured to expose the bases on both strands and we label one of the strands with label.
- This can be a chemiluminescent, radioactive or fluorescent label.

how to prepare the sample for the DNA probe
- extract the double-stranded DNA from the sample and again we denature the DNA to single strands -heat denature the DNA, pulling apart the two strands.

hybridisation reaction in DNA probe use
mix the probe with the DNA from the sample
the probe will then bind to its complementary sequence of DNA within the clinical sample if that particular bacterial species is present within the sample.
remove any non-binding DNA and then have the labelled DNA probe identified within the sample.
- e.g. if this probe targeted the DNA of Porphyromonas gingivalis and Porphyromonas gingivalis was present within the clinical sample, it would bind to Porphyromonas gingivalis DNA as you can see here.

whole genomic DNA probe
use
back in 80s
we didn’t have any genetic information (or genetic sequences) on different bacterial species
whole genomic probe
breakdown of what they probe is
extract and purify the DNA from the bacterial species that you wish to investigate.
- The DNA is cut into much smaller fragments;
- the genome of a bacterial species is round about 4 million
- then label this soup of bacterial DNA fragments to create a whole genomic probe with the label attached.
major issue with whole genomic probes
extremely non-specific
- there is a lot of cross-reactivity between whole genomic probes for one particular bacterial species,
- will cross react with the DNA from other bacterial species because bacteria of different species share so many gene sequences.
- fairly unreliable due to this cross reactivity.
clone gene probes
To identify bacteria within a clinical specimen these cloned gene probes would be prepared targeting a particular gene that might be unique to that particular bacterial species that you wish to identify within the clinical specimen.
- So the gene of interest to be used as a probe would be cloned into E. coli, the cloned fragment isolated, purified and LABEL attached.

clone gene probes Vs whole genomic probes
Now cloned gene probes are much more specific than whole genomic probes.
- In a whole genomic probe we have thousands of genes that have all been labelled,
- whereas in a cloned gene probe we are now dealing with a single gene that has been labelled and ultimately that gene is ideally specific to the bacterial species that you are looking for within the clinical specimen,
therefore the specificity is far higher
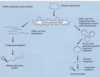
oligonucleotide probes
target
generally target the 16S ribosomal RNA gene, which all bacteria possess.
16S ribosomal RNA (rRNA) gene
all bacteria possess.
essential for survival
gene sequenced for all known bacteria, in public access database
- Approximately 1500 base pairs in length.
- ideal for preparing species-specific probes since it possesses 9 hypervariable regions (V1 to V9)
- contain unique DNA sequences that provide a specific signature for each bacterial species.
- species specific probes or primers for PCR
- contain unique DNA sequences that provide a specific signature for each bacterial species.
- conserved regions - ‘broad range’ (consensus) probes or PCR primers
- Outside the 9 hypervariable regions of gene sequence, the DNA sequence is virtually identical across all bacteria
- can be used to detect bacteria in samples where it is not essential to know the exact species present, but merely if bacteria generally are present or absent
benefit of 16S ribosomal RNA gene oligonucleotide probes
As the gene has been sequenced for all known bacteria, possible to synthesise species-specific probes that target 1 or more of the hypervariable regions.
Synthesised single-stranded oligonucleotide is then labelled and used as previously described.
Oligonucleotides are the DNA probes of choice due to their high specificity due to their small size

hybridisation results

, DNA that had been extracted from subgingival plaque of patients with periodontitis was spotted onto a nylon membrane, the DNA immobilised and subjected to hybridisation with an oligonucleotide probe specific for Porphyromonas gingivalis.
- Hybridisation of the probe has occurred where a black spot is evident, indicating that Porphyromonas gingivalis is present in that sample.
The greater the hybridisation signal (i.e. the larger the black spot) the greater the amount of Porphyromonas gingivalis DNA that is present in that sample.

benefit of 16S rRNA variable regions
9 (V1 to V9)
- Highly variable regions provide unique signatures to any bacterium -> species-specific probes or primers for PCR
benefit of conserved regions in 16S rRNA
“broad range” (consensus) probes or PCR primers
- Outside the nine hypervariable regions of gene sequence, the DNA sequence is virtually identical across all bacteria
- can be used to detect bacteria in samples where it is not essential to know the exact species present, but merely if bacteria generally are present or absent
Polymerase chain reaction
use
Can be used to directly detect bacteria in clinical specimens
PCR
sensitivity and specificity
highly specific and sensitve
what is PCR
exponential amplification procedure whereby a single molecule of DNA can be amplified many billions of times to produce large amounts of DNA.

PCR pratically used in
huge value in forensics, can also be used to detect bacterial within clinical specimens
PCR requires
(4)
- a double stranded DNA template (from the sample),
- primers (specific for a particular gene, for example the 16S ribosomal RNA gene of the bacterial species one wishes to detect in a clinical sample),
- deoxynucleoside triphosphates (dNTPs; the building blocks of DNA)
- the enzyme Taq DNA polymerase - which catalyses the synthesis of new DNA strands.

steps in PCR
- Firstly, the double stranded DNA from the sample is heat denatured at 94oC to pull the 2 DNA strands apart. DENATURATION STEP.
- The PCR primers then hybridise to their target sequences on each DNA strand. PRIMER ANNEALING STEP.
- The Taq DNA polymerase then synthesises the opposite strand, incorporating the dNTPs. PRIMER EXTENSION STEP.
Thus, after each cycle of PCR amplification, the amount of DNA is doubled.
The cycle is then repeated up to 35 times, so after 35 cycles of PCR amplification there has been a 235 amplification of the original DNA.

PCR primers generally target
16S rRNA gene
3 types of PCR primers
- general bacterial primers
- group-specific primers
- species-speicifc primers
general bacterial primers in PCR
- Target the conserves (consensus) region of the 16S ribosomal RNA gene
group-specific primers for PCR
- Detect all members of a particular bacterial genus e.g. Porphyromonas
species specific primers for PCR
2 types
single primer pair (detect a single bacterial species)
more than one primer pair (multiplex)
- detect more than one species at a time
benefit multiplex species spcieif primers
saves time and money
detect more than one species at a time
how many species can generally be ID in PCR with species specific primers at 1 time
usually no more than 3 primer pairs can be used in a single reaction
- since different primer pairs may have different optimal PCR reaction conditions e.g. different primer pairs tend to have different optimum temperatures for primer annealing
how are products of PCR reaction dealt with
separated on an agarose gel which is stained with ethidium bromide and viewed under ultraviolet light.

how to read a PCR reaction on agragose gel
PCR reaction utilised a single primer pair specific for Porphyromonas gingivalis and attempted to detect Porphyromonas gingivalis DNA (and hence the bacterium) in subgingival plaque samples from patients with periodontitis.

- The lanes in which a single PCR product (DNA fragment) is evident indicate which subgingival plaque samples contain Porphyromonas gingivalis.
- The lanes with no PCR product do not contain Porphyromonas gingivalis.
- The larger the amount of the PCR product in each lane, then the greater the amount of Porphyromonas gingivalis in the sample

how to read the results of a multiplex PCR sample
3 primer pairs were used in a single reaction, for detecting 3 different periodontal pathogens (Prevotella intermedia, Treponema denticola and Porphyromonas gingivalis).

The PCR primer pair for each species will yield a PCR product of known size, which will inform us of the bacterial species present in each clinical sample of subgingival plaque from patients with periodontitis.
The leftmost lane contains a calibrated DNA size marker, which allows the sizes of the PCR products to be estimated.
- The first sample contains only one of the bacterial species (Prevotella intermedia)
- the second sample contains two bacterial species (Prevotella intermedia and Treponema denticola),
- the third sample contains only Porphyromonas gingivalis
- The samples in lanes 4 and 8 contain none of these bacteria,
- the sample in lane 9 contains all three bacteria.
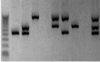
role of calibrated DNA size marker on reading PCR results

allows the sizes of the PCR products to be estimated
(left)

5 advantages of DNA probes and PCR compared to traditional microbiological culture methods for detecting bacteria in clincal samples
- Less time-consuming than culture methods
* Results can be obtained same day for PCR and 24-48hours for DNA probes in general, whereas 7-10 days for culture methods - Very sensitive (DNA probes, 103 cells; PCR, 10 cells; culture methods are 105)
- Can directly detect bacterial DNA within clinical samples
- Do not require viable cells, samples do not have to be analysed immediately
- Can detect uncultivable species these cannot be grown under lab conditions
2 disadvantages of DNA probes and PCR compared to traditional microbiological culture methods for detecting bacteria in clinical samples
- May detect dead cells risk may not have been actively involved in an infection
- Detect only pre-selected species
- Culture methods can detect a multitude of different bacteria within samples
what is PCR-RFLP
restriction fragment length polymorphism
use of PCR-RFLP
Identification of bacterial isolates, that have been obtained by culture methods
Digest PCR-amplified bacterial 16S rRNA gene with restriction enzymes
- as the 16S ribosomal RNA gene sequence has been determined for all known bacterial species, it is possible to predict the sizes of DNA fragments obtained when digesting the gene with restriction enzyme
Yields specific patterns (“fingerprints”) for individual species – ID near 100% accuracy
Rapid, cheaper, more specific alternative to biochemical tests for identifying bacterial isolates from clinical samples
benefits of PCR-RFLP
Rapid, cheaper, more specific alternative to biochemical tests for identifying bacterial isolates from clinical samples
end product of PCR-RFLP
- Yields specific patterns (“fingerprints”) for individual species – ID near 100% accuracy
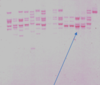
how to analyse outcome of PCR-RFLP

The DNA fragments obtained from digesting the 16S rRNA gene of each bacterial isolate have been separated on an agarose gel.
Different bacterial species have different patterns.
- For example, blue arrow 4 isolates are the same species as they possess the same collection (or pattern or fingerprint) of DNA fragments, and they in turn are different from other isolates

sufficient genetic diversity exits which allows
identification of different clones (strains)among isolates of the same species
- subtyping
2 reasons why it is important to subtype bacteria
- Track routes of transmission during disease outbreaks
- Study pathogenicity of specific strains
traditional methods of subtyping bacteria (2)
Testing for one or more phenotypic markers:
- Serotyping
- Biotyping
3 problems with traditional methods of subtyping bacteria
- Limited discriminatory capacity not effective at distinguishing between different strain of the same species
- Organism-specific methods
- Specialised reagents required
examples of molecular (genetic) typing methods (5)
restriction enzyme analysis (REA)
gene probe typing
- ribotyping
16S-23S intergenic spacer region (IGS) - PCR-RFLP of it
DNA sequencing
restriction enzyme analysis
type of molecular (genetic) typing
- Digest whole genomic DNA with restriction enzymes
probelm with REA
Problem
- Too many DNA fragments obtained, makes interpretation difficult (cannot be resolved in agarose gel)
example of REA
The DNA fragments tend to merge into each other, producing a smear effect.
- Because of this, it is almost impossible to discriminate between different strains.

gene probe typing benefit
Can reduce number of DNA fragments generated by REA using a suitable gene probe

ribotyping is
type of gene probe typing
Use E. coli rRNA operon (16S-23S-5S) as a DNA probe following REA
- rRNA operon present in multiple copies in bacterial genomes
- well conserved in overall structure and sequence due to its evolutionary role in all bacteria
- Variation in number and size of fragments in bacterial DNA digests which are complementary with rRNA
ribotyping uses
- E. coli ribosomal RNA operon (16S-23S-5S) as a DNA probe following REA*
- is comprised of adjoining 16S, 23S and 5S ribososmal RNA genes
process of ribotyping (gene probe typing)
Genomic DNA is extracted from the bacterial isolates or clinical sample and digested with 1 or more restriction enzymes to give many DNA fragments.
- DNA fragments are separated into distinct bands by agarose gel electrophoresis
- DNA bands in the gel are transferred to a nylon membrane in preparation for hybridisation to the labelled DNA probe
- for ribotyping, the probe is the E. coli ribosomal RNA operon
- The DNA probe binds to specific DNA fragments on the membrane that all or part of the probe sequence.
- The probe is then visualised and the DNA fragments compared.

rule of agarose gel electrophoresis movement
the smaller more mobile DNA fragments migrate more quickly than larger DNA fragments.
how to analyse outcome of ribotyping different bacterial strains

Distinct DNA “fingerprints” are clearly visible between different strains.
- For example, these 2 strains are the same as they share an identical DNA “fingerprint” but are different from other strains which have different DNA “fingerprints”.
16S-23S intergenic spacer region (IGS) typing
Very variable sequence
Amplify this region by PCR using consensus primers
Digest PCR products with 1 or more restriction enzymes to obtain strain-specific DNA fingerprints
The intergenic spacer region that lies between the 16S and 23S ribosomal RNA genes is amplified by PCR using primers as shown

16S-23S intergenic spacer region (IGS)
very variable sequence
- Even different strains of the same bacterial species will have a different sequences and restriction enzyme profile i.e. different numbers of sites exists at which any enzyme will cut the DNA
16S-23S intergenic spacer region (IGS) typing benefit
very rapid and technically simpler DNA-based typing method is PCR-RFLP analysis of the 16S-23S intergenic spacer region
compared to ribotyping
how to analyse PCR-RFLP of 16S-23S intergenic spacer of different strains of 1 bacterial species
- Distinct DNA fingerprints can be seen between different strains.
For example, these 3 strains are the same as they share an identical DNA “fingerprint” but are different from other strains which have different DNA “fingerprints”.

DNA sequencing as typing methods
ultimate
- Can detect single base differences between strains
sequences all the bases in a stretch of DNA and detects even single base changes between different bacterial strains of the same species.
how to read a sanger sequence reaction output
read from the bottom of the gel upwards by counting the order in which the bases in the A, C, G and T lanes appear: C, A, T, G, C, C etc

molecular identification of uncutivable and novel bacteria
Firstly genomic DNA is extracted from the clinical sample.
General bacterial PCR primers are then used to amplify the 16S ribosomal RNA gene from all the bacteria in the clinical sample.
- if 200 bacteria are present in the sample -> 200 different bacterial 16S ribosomal RNA genes will be amplified.
Since the 16S ribosomal RNA genes are of a very similar size (approximately 1500 base pairs in length), they must be separated by cloning into a suitable plasmid vector and then introduced into E. Coli.
From the E. Coli library generated, 50 clones are randomly chosen and their 16S ribosomal RNA genes sequenced using the Sanger method.
- representative snapshot of the clone library.
Once the clone sequences are obtained, the 16S ribosomal RNA genes of the unknown bacterial species in the samples are analysed using a programme called BLAST
- compares their 16S ribosomal RNA gene sequences to those of known bacterial species in public access databases.

if an uncutivable/novel bacteria species has a gene sequence match of 98% or higher with a known 16S ribosomal RNA gene using BLAST
the clone sequence as having originated from that particular bacterial species, which was therefore present in the sample.
if an uncultivable/novel bacteria’s gene sequence match is less than 97% with known gene sequences in the public database indicates that
that a potentially novel bacterial species has been identified in the clinical sample.


