Ch1 Flashcards
What are the six distinguishing features of living organisms?
A high degree of chemical complexity and microscopic organization.
Systems for extracting, transforming, and using energy from the environment.
Defined functions for each of an organism’s components and regulated interactions among them.
Mechanisms for sensing and responding to alterations in their surroundings.
A capacity for precise self-replication and self-assembly.
A capacity to change over time by gradual evolution.
What boundry does the plasma membrane define?
The perhiphery of the cell, separating the contents from the surroundings.
What are the universal features of living cells?
A nucleous or nucleoid, a plasma membrane, cytoplasm.

The cytosol is defined as what portion of the cytoplasm?
The cytosol is defined as that portion of the cytoplasm that remains in the supernatant after gentle breakage of the plasma membrane and centrifugation of the resulting extract at 150,000 g for 1 hour.

What remains in the supernatant of cytoplasm centrifuged at 150,000 g for one hour?
The cytosol, the supernatant of cytoplasm, a concentrated solution of enzymes, RNA, monomeric subunits, metabolites, and inorganic ions.

What forms the pellet of cytosol when centrifuged at 150,000 g for one hour?
After the cytosol (the supernatant) is removed, particles and organelles are what remains:
Ribosomes, storage granules, mitochondria, chloroplasts, lysosomes, endoplasmic reticulum.

Define the two types of phototrophs by carbon source and give examples of each:
Autotrophs: carbon from CO2 (inorganic).
ex: cyanobacteria, vascular plants
Heterotrophs: carbon from organic compounds.
ex: purple bacteria, green bacteria

Define the two types of chemotrophs by energy source and give examples of each:
Lithotrophs: oxidise inorganic fuels.
ex: sulfur bacteria, hydrogen bacteria
Organotrophs: oxidise organic fuels.
ex: most bacteria, all nonphototrophic eukaryotes
Both types may be either autotropic or hetertrophic with regard to carbon source used for catabolism.

List common features of bacterial cells:
Nucleoid, ribosomes, pili, flagella, cell envelope (Gram - or Gram +)

ribosome
ribosomes synthesise protein from an RNA message, 70S in bacteria, 80S in eukaryotes.

nucleoid
the nucleoid contains a single, simple, long circular DNA molecule, not membrane bound

pili
pili provide points of adhesion to surface of other cells.

flagella
flagellum are used propel cell through its surroundings.

Gram + vs Gram -
Gram - : Inner membrane, thinner (relative to Gram +) peptidoglycan layer, LPS (lippopolysaccharide) outer membrane.
Gram + : Inner membrane, thinner (relative to Gram -) peptidoglycan layer, no outer membrane.

peroxisome
peroxisome is a vesicle present in cytoplasm that oxidises fatty acids

cytoskeleton
cytoskeleton supports the cell, aids in movement of organelles

nucelar envelope
nucelar envelope segregates chromatin (DNA protein) from cytoplasm

lysosome
lysosome is a vesicle present in cytoplasm that degrades intracellular debris (animal cells only)

Golgi complex
Golgi complex processes, packages, and targets proteins to other organelles or for export

smooth endoplasmic reticulum
smooth endoplasmic reticulum is the site of lipid synthesis and drug metabolism

rough endoplasmic reticulum
rough endoplasmic reticulum is the site of much protein synthesis

mitochondrion
mitochondrion oxidises fuels to produce ATP

transport vesicle
transport vesicle shuttles lipids and proteins between the endoplasmic recticulum, Golgi complex, and plasma membrane

nucleolus
nucleolus is the site of ribosomal RNA (rRNA) synthesis

proteasome
Proteasomes are protein complexes inside all eukaryotes and archaea, and in some bacteria. In eukaryotes, they are located in the nucleus and the cytoplasm. The main function of the proteasome is to degrade unneeded or damaged proteins by proteolysis, a chemical reaction that breaks peptide bonds.
chloroplast
chloroplast harvests sunlight, produces ATP and carbohydrates (plant cells only)
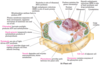
starch granule
starch granules temporarily store carbohydrate products of photosynthesis (plant cells only)
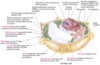
thylakoids
thylakoids are the site of light driven ATP synthesis (plant cells only)

cell wall
cell wall provides shape and rigidity; protects cell from osmotic swelling (plant cells only)
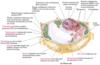
vacuole
vacuoles degrade and recycle macromolecules, stores metabolites (plant cells only)

plasmodesma
opening in the cell wall that provides a path between two plant cells (plant cells only)

glyoxysome
glyoxysomes contain enzymes of the glyoxylate cycle (plant cells only)

describe the contents of the pellet that results from centrifugation of a homogenised tissue sample at 1,000 g for 10 minutes

describe the process of differental centrifugation
Differential centrifugation is a common procedure in microbiology and cytology used to separate certain organelles from whole cells for further analysis of specific parts of cells. In the process, a tissue sample is first homogenised to break the cell membranes and mix up the cell contents. The homogenate is then subjected to repeated centrifugations, each time removing the pellet and increasing the centrifugal force. Finally, purification may be done through equilibrium sedimentation, and the desired layer is extracted for further analysis.

What is the typical size of an animal or plant cell?
5 to 100 𝛍m in diameter
supernatant
The liquid layer remaining after centrifugation, contrast with the pellet (precipitate in diagram shown)

Isopycnic centrifugation is effective for separating what kinds of particles?
Those of a different density.
In isopycnic centrifugation, a centrifuge tube is filled with a solution, the density of which increases from top to bottom; a solute such as sucrose is dissolved at different concentrations to produce the density gradient. When a mixture of organelles is layered on top of the density gradient and the tube is centrifuged at high speed, individual organelles sediment until their buoyant density exactly matches that in the gradient. Each layer can be collected separately

isopycnic centrifugation
In isopycnic centrifugation, a centrifuge tube is filled with a solution, the density of which increases from top to bottom; a solute such as sucrose is dissolved at different concentrations to produce the density gradient. When a mixture of organelles is layered on top of the density gradient and the tube is centrifuged at high speed, individual organelles sediment until their buoyant density exactly matches that in the gradient. Each layer can be collected separately

What is the size of a typical unicellular microorganism?
1 to 2 𝛍m long
what are the three general types of cytoskeleton filaments?
actin filaments, microtubules, intermediate filaments
(shown: Mitosis in a newt lung cell. Microtubules (green), attached to structures called kinetochores (yellow) on the condensed chromosomes (blue), pull the chromosomes to opposite poles, or centrosomes (magenta), of the cell. Intermediate filaments, made of keratin (red), maintain the structure of the cell)

What is the main difference between prokaryotic and eukaryotic ribosomes?
Prokaryotic ribosomes are smaller (70S vs 80S) than their eukaryotic counterparts.
name nucleotide U
Uracil

name nucleotide T
Thymine, 2 bonds with Adenine

name nucleotide C
Cytosine

name nucleotide A
Adenine

name nucleotide G
Guanine

what nitrogenous base is this? what type of molecule is it? where is it attatched to the ribose backbone and how many hydrogen bonds does it form in standard Watson-Crick configuration? If it can be commonly methylated, where?

Thymine, a pyrimidine, is connected to the ribose backbone at position 1, and forms two hydrogen bonds with Adenine at positions 3,4.
IUPAC name for position clarity: 5-Methylpyrimidine-2,4(1H,3H)-dione

what nitrogenous base is this? what type of molecule is it? where is it attatched to the ribose backbone and how many hydrogen bonds does it form in standard Watson-Crick configuration? If it can be commonly methylated, where?

Guanine, a purine, is connected to the ribose backbone at position 9 and forms three hydrogen bonds with Cytosine.
IUPAC name for position clarity: 2-amino-9H-purin-6(1H)-one

what nitrogenous base is this? what type of molecule is it? where is it attatched to the ribose backbone and how many hydrogen bonds does it form in standard Watson-Crick configuration? If it can be commonly methylated, where?

Cytosine, a pyrimidine, is connected to the ribose backbone at position 1, and forms three hydrogen bonds to Guanine. Can be methylated at position 5.
IUPAC name for position clarity: 4-aminopyrimidin-2(1H)-one

what nitrogenous base is this? what type of molecule is it? where is it attatched to the ribose backbone and how many hydrogen bonds does it form in standard Watson-Crick configuration? If it can be commonly methylated, where?

Adenine, a purine, is connected to the ribose backbsone at position 3 and forms two hydrogen bonds with Guanine.
IUPAC name for position clarity: 9H-purin-6-amine

What four elements are the most abundant in living organisms and why?
The four most abundant elements in living organisms, in terms of percentage of total number of atoms, are hydrogen, oxygen, nitrogen, and carbon, which together make up more than 99% of the mass of most cells. They are the lightest elements capable of forming one, two, three, and four bonds, respectively; in general, the lightest elements form the strongest bonds.

methyl


methyl
amino


amino
ethyl


ethyl
amido


amido
phenyl


phenyl
guanidino
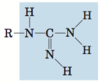

guanidino
carbonyl

aldehyde
carbonyl


carbonyl
(aldehyde)
imidazole


imidazole
ketone
carbonyl


carbonyl
(ketone)
sulfhydryl


sulfhydryl
disulfide


disulfide
carboxyl

hydroxyl

ether

ester

thioester

phosphoryl

phosphoanhydride

mixed anhydride
(shown: dehydration of carboxylic acid and phosphoric acid, also called acyl phosphate)

anhydride
(dehydration of two carboxcylic acids)


carboxyl

hydroxyl

ether

ester

thioester

phosphoryl

phosphoanhydride

mixed anhydride

anhydride
acetyl


acetyl
Vitamin B5
Pantothenic acid, also called pantothenate or vitamin B5, is a water-soluble vitamin. For many animals, pantothenic acid is an essential nutrient. Animals require pantothenic acid to synthesize coenzyme-A (CoA), as well as to synthesize and metabolize proteins, carbohydrates, and fats.

pantothenic acid
Pantothenic acid, also called pantothenate or vitamin B5, is a water-soluble vitamin. For many animals, pantothenic acid is an essential nutrient. Animals require pantothenic acid to synthesize coenzyme-A (CoA), as well as to synthesize and metabolize proteins, carbohydrates, and fats.

describe ATP
Adenosine triphosphate (ATP)
The removal of the terminal phosphoryl group (shaded pink) of ATP, by breakage of a phosphoanhydride bond, is highly exergonic, and this reaction is coupled to many endergonic reactions in the cell

nucleus
nucleus contains the genes (chromatin)
what organises the cytoplasm?
cytoskeleton
describe cytoskeletal filaments and their structure:
Each type of cytoskeletal component is composed of simple protein subunits that associate noncovalently to form filaments of uniform thickness. These filaments are not permanent structures; they undergo constant disassembly into their protein subunits and reassembly into filaments. Their locations in cells are not rigidly fixed but may change dramatically with mitosis, cytoki- nesis, amoeboid motion, or changes in cell shape. The assembly, disassembly, and location of all types of fila- ments are regulated by other proteins, which serve to link or bundle the filaments or to move cytoplasmic organelles along the filaments.
what is the purpose of the endomembrane system?
segregates specific metabolic processes and provides surfaces on which certain enzyme-catalysed reactions occur
exocytosis
endocytosis
transport into a cell
Generally, why are trace elements essential for life even though they are present in such small relative quantaties?
Usually because they are essential for the function of specific proteins, including those that act as enzymes.
Describe the bond angles and length of a typical carbon atom bound to four ligands.
109.5° with an average length of .154 nm
What is a polyfunctional molecule?
A biomolecule with two or more different kinds of functional groups
what is dissolved in the cytosol of all cells?
100 to 200 central metabolites of the major pathways that occur in nearly every cell (Mr ~100 to ~500), including the common amino acids, nucleotides, sugars and their phosphorylated derivatives, and a number of mono-, di-, and tricarboxylic acids.
secondary metabolites
Secondary metabolites are organic compounds that are not directly involved in the normal growth, development, or reproduction of an organism. Unlike primary metabolites, absence of secondary metabolites does not result in immediate death, but rather in long-term impairment of the organism’s survivability, fecundity, or aesthetics, or perhaps in no significant change at all. Secondary metabolites are often restricted to a narrow set of species within a phylogenetic group. Secondary metabolites often play an important role in plant defense against herbivory and other interspecies defenses. Humans use secondary metabolites as medicines, flavorings, and recreational drugs.
primary metabolites
A primary metabolite is a kind of metabolite that is directly involved in normal growth, development, and reproduction. It usually performs a physiological function in the organism (i.e. an intrinsic function).
A primary metabolite is typically present in many organisms or cell, while a central metabolite has an even more restricted meaning: present in any autonomously growing cell or organism
primary metabolite vs. central metabolite
A primary metabolite is typically present in many organisms or cell, while a central metabolite has an even more restricted meaning: present in any autonomously growing cell or organism
metabolome
the entire collection of small molecules (metabolites) in a given cell, similar to a cell’s genome or proteome
Mr
Molecular weight, or relative molecular mass, denoted Mr. The molecular weight of a substance is defined as the ratio of the mass of a molecule of that substance to one-twelfth the mass of carbon-12 (12C). Since Mr is a ratio, it is dimensionless—it has no associated units. The second is molecular mass, denoted m. This is simply the mass of one molecule, or the molar mass divided by Avogadro’s number. The molecular mass, m, is expressed in daltons (abbreviated Da). One dalton is equivalent to one-twelfth the mass of carbon-12; a kilodalton (kDa) is 1,000 daltons; a megadalton (MDa) is 1 million daltons.
Consider, for example, a molecule with a mass 1,000 times that of water. We can say of this molecule either Mr = 18,000 or m = 18,000 daltons. We can also describe it as an “18 kDa molecule.” However, the expression Mr = 18,000 daltons is incorrect, because Mr is dimensonless.
Another convenient unit for describing the mass of a single atom or molecule is the atomic mass unit (formerly amu, now commonly denoted u). One atomic mass unit (1 u) is defined as one-twelfth the mass of an atom of carbon-12. Since the experimen- tally measured mass of an atom of carbon-12 is 1.9926 x 10-23 g, 1 u = 1.6606 x 10-24 g. The atomic mass unit is convenient for describing the mass of a peak observed by mass spectrometry.
numerically, Mr = Da = u
define closed and open systems in relation to the universe
For chemical reactions occurring in solution, we can define a system as all the reactants and products present, the solvent that contains them, and the immediate atmosphere—in short, everything within a defined region of space. The system and its surroundings together constitute the universe. If the system exchanges neither matter nor energy with its surroundings, it is said to be isolated. If the system exchanges energy but not matter with its surroundings, it is a closed system; if it exchanges both energy and matter with its surroundings, it is an open system.
living organisms are isolated, open or closed systems?
open systems, they exchange energy and matter with their surroundings
first law of thermodynamics
in any physical or chemical change, the total amount of energy in the universe remains constant, although the form of the energy may change.
how is the randomness or disorder of the components of a system expressed?
entropy, S
any change in the randomness of the system is expressed as entropy change, ΔS, by convention a positive value when randomness increases
what is meant by a positive ΔS ?
an increase in randomness of the system
what is the formula for Gibbs free energy?
G = H - TS
free-energy content, G, of any closed system can be defined in terms of three quantities: enthalpy, H, reflecting the number and kinds of bonds; entropy, S; and the absolute temperature, T (in degrees Kelvin)
at what value of Gibbs free-energy change do reactions tend to occur spontaneously?
at negative ΔG
ΔG°
The standard free-energy change for a reaction, ΔG°, is a physical constant that is related to
the equilibrium constant by the equation:
ΔG° = -RT ln Keq.


